2024-02-07 イリノイ大学アーバナ・シャンペーン校
<関連情報>
- https://aces.illinois.edu/news/new-study-finds-corn-genome-can-gang-multiple-pathogens-once
- https://academic.oup.com/g3journal/article/doi/10.1093/g3journal/jkad275/7459157?login=false
トウモロコシの4つの葉面病に対する抵抗性遺伝子座の同定 Identification of loci conferring resistance to 4 foliar diseases of maize
Yuting Qiu, Pragya Adhikari, Peter Balint-Kurti, Tiffany Jamann
G3 Genes|Genomes|Genetics Published:05 December 2023
DOI:https://doi.org/10.1093/g3journal/jkad275
Abstract
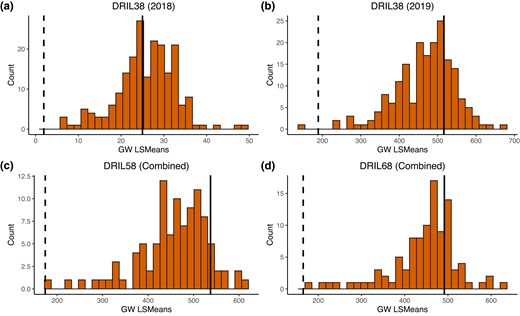
Foliar diseases of maize are among the most important diseases of maize worldwide. This study focused on 4 major foliar diseases of maize: Goss’s wilt, gray leaf spot, northern corn leaf blight, and southern corn leaf blight. QTL mapping for resistance to Goss’s wilt was conducted in 4 disease resistance introgression line populations with Oh7B as the common recurrent parent and Ki3, NC262, NC304, and NC344 as recurrent donor parents. Mapping results for Goss’s wilt resistance were combined with previous studies for gray leaf spot, northern corn leaf blight, and southern corn leaf blight resistance in the same 4 populations. We conducted (1) individual linkage mapping analysis to identify QTL specific to each disease and population; (2) Mahalanobis distance analysis to identify putative multiple disease resistance regions for each population; and 3) joint linkage mapping to identify QTL across the 4 populations for each disease. We identified 3 lines that were resistant to all 4 diseases. We mapped 13 Goss’s wilt QTLs in the individual populations and an additional 6 using joint linkage mapping. All Goss’s wilt QTL had small effects, confirming that resistance to Goss’s wilt is highly quantitative. We report several potentially important chromosomal bins associated with multiple disease resistance including 1.02, 1.03, 3.04, 4.06, 4.08, and 9.03. Together, these findings indicate that disease QTL distribution is not random and that there are locations in the genome that confer resistance to multiple diseases. Furthermore, resistance to bacterial and fungal diseases is not entirely distinct, and we identified lines resistant to both fungi and bacteria, as well as loci that confer resistance to both bacterial and fungal diseases.