次世代材料創製の新たな可能性を切り開く画期的な成果 Breakthrough Opens Up New Possibilities for Creation of Next Generation of Materials
2022-09-28 ニューヨーク大学 (NYU)
エマルション(水に浸した油滴)を利用して、フォルダーの自己組織化を行うもので、フォルダーとは、液滴の一連の相互作用から理論的に予測できるユニークな形状のことである。
この自己組織化プロセスは、コロイドを用いたタンパク質やRNAの折り畳みを模倣した生物学の分野から引用したものである。Nature誌掲載の研究では、水中に油性の微小な液滴を作り、そこに組み立ての「指示書」となるDNA配列を持たせた。この液滴は、まず柔軟な鎖に集合し、次に粘着性のあるDNA分子を介して順次折りたたまれていく。この折りたたみにより、12種類のフォルダーが生成され、さらに特異性を高めると、600種類ある幾何学的形状の半分以上をコード化することができる。
<関連情報>
- https://www.nyu.edu/about/news-publications/news/2022/september/physicists-take-self-assembly-to-new-level-by-mimicking-biology.html
- https://www.nature.com/articles/s41586-022-05198-8
プログラマブルフォールディングによるエマルション液滴の自己組織化 Self-assembly of emulsion droplets through programmable folding
Angus McMullen,Maitane Muñoz Basagoiti,Zorana Zeravcic & Jasna Brujic
Nature Published:28 September 2022
DOI:https://doi.org/10.1038/s41586-022-05198-8
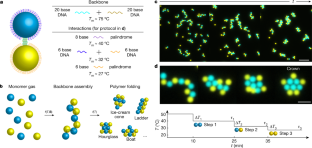
Abstract
In the realm of particle self-assembly, it is possible to reliably construct nearly arbitrary structures if all the pieces are distinct1,2,3, but systems with fewer flavours of building blocks have so far been limited to the assembly of exotic crystals4,5,6. Here we introduce a minimal model system of colloidal droplet chains7, with programmable DNA interactions that guide their downhill folding into specific geometries. Droplets are observed in real space and time, unravelling the rules of folding. Combining experiments, simulations and theory, we show that controlling the order in which interactions are switched on directs folding into unique structures, which we call colloidal foldamers8. The simplest alternating sequences (ABAB…) of up to 13 droplets yield 11 foldamers in two dimensions and one in three dimensions. Optimizing the droplet sequence and adding an extra flavour uniquely encodes more than half of the 619 possible two-dimensional geometries. Foldamers consisting of at least 13 droplets exhibit open structures with holes, offering porous design. Numerical simulations show that foldamers can further interact to make complex supracolloidal architectures, such as dimers, ribbons and mosaics. Our results are independent of the dynamics and therefore apply to polymeric materials with hierarchical interactions on all length scales, from organic molecules all the way to Rubik’s Snakes. This toolbox enables the encoding of large-scale design into sequences of short polymers, placing folding at the forefront of materials self-assembly.



