2025-09-29 ペンシルベニア州立大学(Penn State)
<関連情報>
- https://www.psu.edu/news/agricultural-sciences/story/new-clues-how-plant-microbiomes-protect-against-bacterial-speck-disease
- https://link.springer.com/article/10.1186/s40793-025-00734-1
実験的マイクロバイオーム選択後のトマトにおける病害抑制性葉圏マイクロバイオームの迅速かつ持続的な分化 Rapid and sustained differentiation of disease-suppressive phyllosphere microbiomes in tomato following experimental microbiome selection
Hanareia Ehau-Taumaunu,Terrence H. Bell,Javad Sadeghi & Kevin L. Hockett
Environmental Microbiome Published:01 July 2025
DOI:https://doi.org/10.1186/s40793-025-00734-1
Abstract
Background
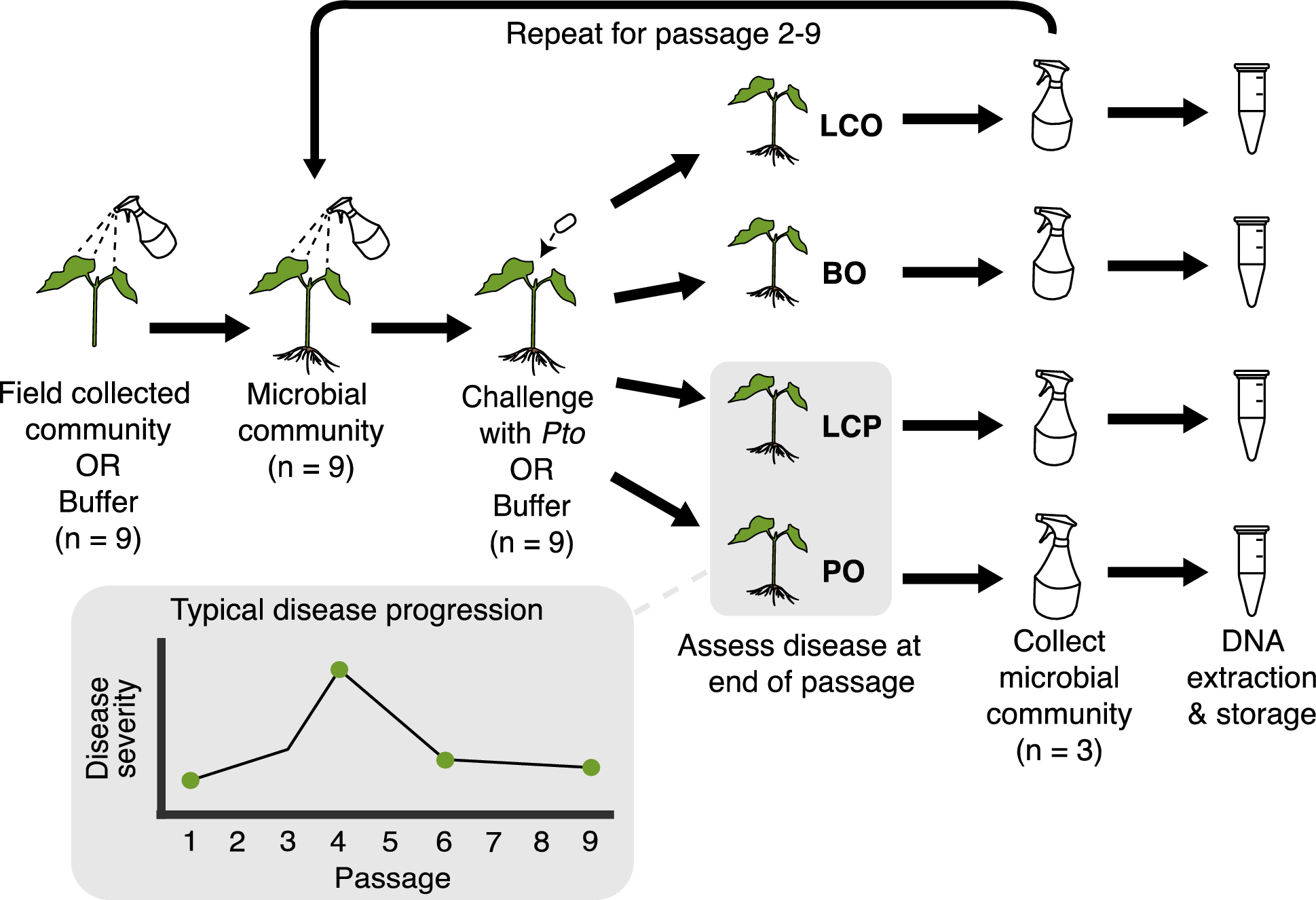
Microbial-based treatments to protect plants against phytopathogens typically focus on soil-borne disease or the aboveground application of one or a few biocontrol microorganisms. However, diverse microbiomes may provide unique benefits to phytoprotection in the phyllosphere, by restricting pathogen access to niche space and/or through multiple forms of direct antagonism. We previously showed that successive experimental passaging of phyllosphere microbiomes along with the phytopathogen Pseudomonas syringae pv. tomato (Pto), which causes bacterial speck in tomato, led to the development of a disease suppressive microbial community. Here, we used amplicon sequencing to assess bacterial and fungal composition at the end of each passage, as well as shotgun metagenomics at key passages based on observed disease-suppressive phenotypes, to assess differences in functional potential between suppressive and non-suppressive communities.
Results
Bacterial composition changed and diversity declined quickly due to passaging and remained low, particularly in treatments with Pto present, whereas fungal diversity did not. Pseudomonas and Xanthomonas populations were particularily enriched in disease-suppressive microbiomes compared to conducive microbiomes. The relative abundance of Pseudomonas syringae group gemonosp. 3 (the clade to which the introduced pathogen belongs) in shotgun metagenomic data was similar to what we observed for Pseudomonas ASVs in the 16S rRNA gene dataset. We also observed an increase in the abundance of genes associated with microbial antagonism at Passage 4, corresponding to the highest observed disease severity.
Conclusions
Taxonomic richness and evenness were low within samples, with clustering occurring for suppressive or non-suppressive microbiomes. The relative abundance of genes associated with antagonism was higher for disease-suppressive phyllosphere microbiomes. This work is an important step towards understanding the microbe-microbe interactions within disease-suppressive phyllosphere communities.



