2026-01-21 ニューヨーク大学(NYU)

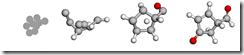
A new AI model designs molecules with specified properties 10 times faster than previous methods, potentially speeding up the process for the creation of pharmaceuticals and materials. The figure illustrates how the system transforms random noise into complete molecular structures guided by target properties. Image courtesy of the University of Florida and New York University
<関連情報>
- https://www.nyu.edu/about/news-publications/news/2026/january/scientists-design-molecules–backward–to-speed-up-discovery.html
- https://www.nature.com/articles/s43588-025-00946-y
PropMolFlow: ジオメトリ完全なフローマッチングによる特性誘導分子生成 PropMolFlow: property-guided molecule generation with geometry-complete flow matching
Cheng Zeng,Jirui Jin,Connor Ambrose,George Karypis,Mark Transtrum,Ellad B. Tadmor,Richard G. Hennig,Adrian Roitberg,Stefano Martiniani & Mingjie Liu
Nature Computational Science Published:21 January 2026
DOI:https://doi.org/10.1038/s43588-025-00946-y
A preprint version of the article is available at arXiv.
Abstract
Molecule generation is advancing rapidly in chemical discovery and drug design. Flow-matching methods have recently set the state of the art (SOTA) in unconditional molecule generation, surpassing score-based diffusion models. However, diffusion models still lead in property-guided generation. In this work, we introduce PropMolFlow, an approach for property-guided molecule generation based on geometry-complete SE(3)-equivariant flow matching. Integrating five different property embedding methods with a Gaussian expansion of scalar properties, PropMolFlow achieves competitive performance against previous SOTA diffusion models in conditional molecule generation while maintaining high structural stability and validity. Additionally, it enables higher sampling speed with fewer time steps compared with baseline models. We highlight the importance of validating the properties of generated molecules through density functional theory calculations. Furthermore, we introduce a task to assess the model’s ability to propose molecules with under-represented property values, assessing its capacity for out-of-distribution generalization.