2025-11-03 英国研究イノベーション機構(UKRI)
Web要約 の発言:
<関連情報>
- https://www.ukri.org/news/new-gene-atlas-paves-way-for-healthier-more-resilient-oats/
- https://www.nature.com/articles/s41586-025-09676-7
- https://www.nature.com/articles/s41467-025-57895-3
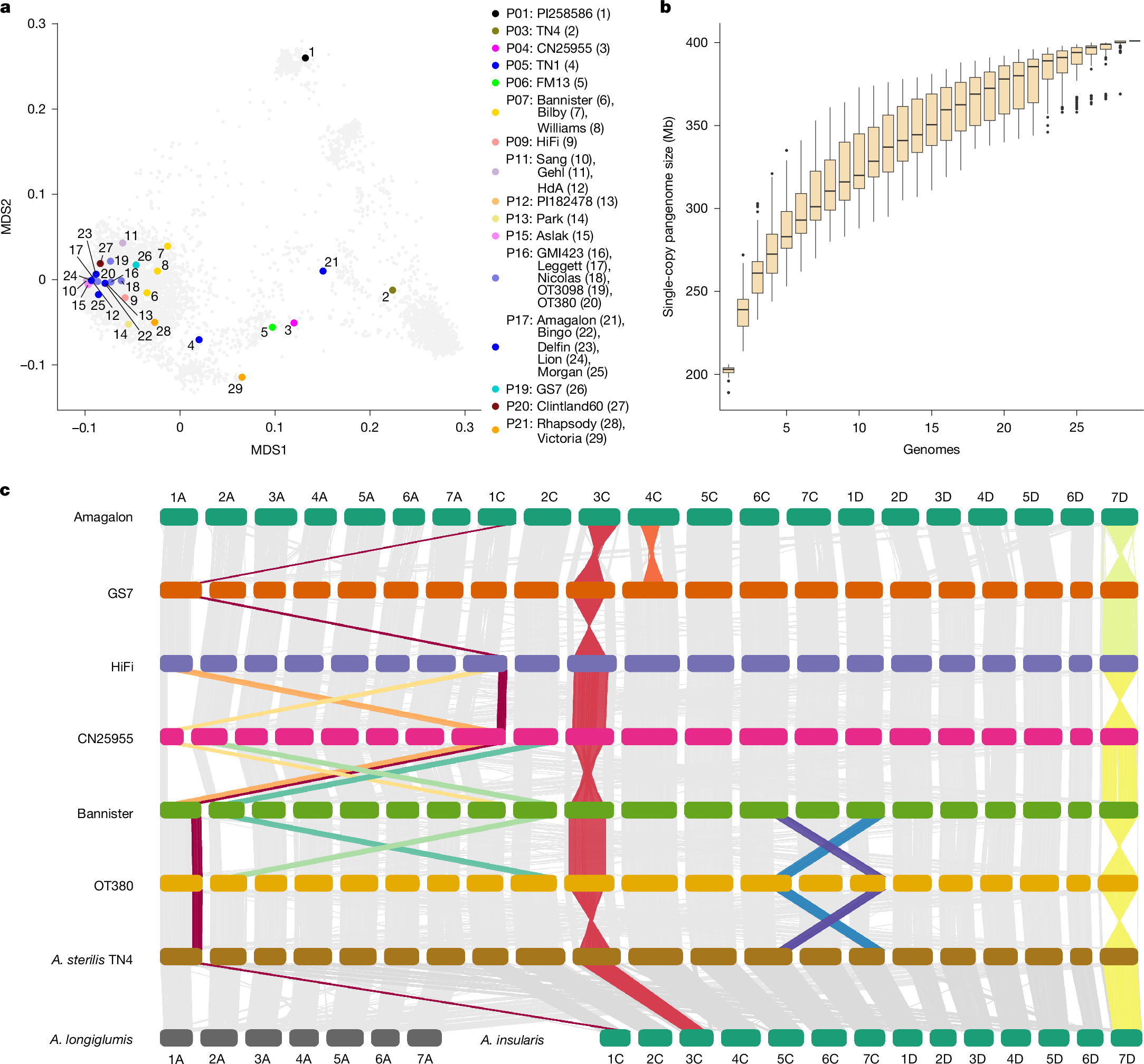
六倍体オート麦のパンゲノムとパントランスクリプトーム A pangenome and pantranscriptome of hexaploid oat
Raz Avni,Nadia Kamal,Lidija Bitz,Eric N. Jellen,Wubishet A. Bekele,Tefera T. Angessa,Petri Auvinen,Oliver Bitz,Brian Boyle,Francisco J. Canales,Craig H. Carlson,Brett Chapman,Harmeet Singh Chawla,Yutang Chen,Dario Copetti,Samara Correia de Lemos,Viet Dang,Steven R. Eichten,Kathy Esvelt Klos,Amit M. Fenn,Anne Fiebig,Yong-Bi Fu,Heidrun Gundlach,Rajeev Gupta,… Martin Mascher
Nature Published:29 October 2025
DOI:https://doi.org/10.1038/s41586-025-09676-7
Abstract
Oat grain is a traditional human food that is rich in dietary fibre and contributes to improved human health1,2. Interest in the crop has surged in recent years owing to its use as the basis for plant-based milk analogues3. Oat is an allohexaploid with a large, repeat-rich genome that was shaped by subgenome exchanges over evolutionary timescales4. In contrast to many other cereal species, genomic research in oat is still at an early stage, and surveys of structural genome diversity and gene expression variability are scarce. Here we present annotated chromosome-scale sequence assemblies of 33 wild and domesticated oat lines, along with an atlas of gene expression across 6 tissues of different developmental stages in 23 of these lines. We construct an atlas of gene-expression diversity across subgenomes, accessions and tissues. Gene loss in the hexaploid is accompanied by compensatory upregulation of the remaining homeologues, but this process is constrained by subgenome divergence. Chromosomal rearrangements have substantially affected recent oat breeding. A large pericentric inversion associated with early flowering explains distorted segregation on chromosome 7D and a homeologous sequence exchange between chromosomes 2A and 2C in a semi-dwarf mutant has risen to prominence in Australian elite varieties. The oat pangenome will promote the adoption of genomic approaches to understanding the evolution and adaptation of domesticated oats and will accelerate their improvement.
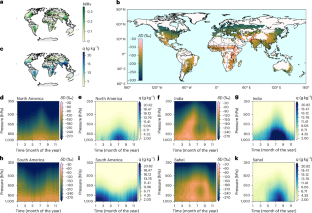
野生および栽培オート麦のゲノム集団構造は染色体再編成の兆候を明らかにする Global genomic population structure of wild and cultivated oat reveals signatures of chromosome rearrangements
Wubishet A. Bekele,Raz Avni,Clayton L. Birkett,Asuka Itaya,Charlene P. Wight,Justin Bellavance,Sophie Brodführer,Francisco J. Canales,Craig H. Carlson,Anne Fiebig,Yongle Li,Steve Michel,Raja Sekhar Nandety,David J. Waring,Juan D. Arbelaez,Aaron D. Beattie,Melanie Caffe,Isabel A. del Blanco,Jason D. Fiedler,Rajeev Gupta,Lucia Gutierrez,John C. Harris,Stephen A. Harrison,Matthias H. Herrmann,… Nicholas A. Tinke
Nature Communications Published:29 October 2025
DOI:https://doi.org/10.1038/s41467-025-57895-3
Abstract
The genus Avena consists of approximately 30 wild and cultivated oat species. Cultivated oat is an important food crop, yet the broader genetic diversity within the Avena gene pool remains underexplored and underexploited. Here, we characterize over 9000 wild and cultivated hexaploid oat accessions of global origin using genotyping-by-sequencing and explore population structure using multidimensional scaling and population-based clustering methods. We also conduct analyses to reveal chromosome regions associated with local adaptation, sometimes resulting from large-scale chromosome rearrangements. We report four distinct genetic populations within the wild species A. sterilis, a distinct population of cultivated A. byzantina, and multiple populations within cultivated A. sativa. Some chromosome regions associated with local adaptation are also associated with confirmed structural rearrangements on chromosomes 1A, 1C, 3C, 4C, and 7D. This work provides evidence suggesting multiple polyploid origins, multiple domestications, and/or reproductive barriers amongst Avena populations caused by differential chromosome structure.