2025-08-18 北京大学(PKU)
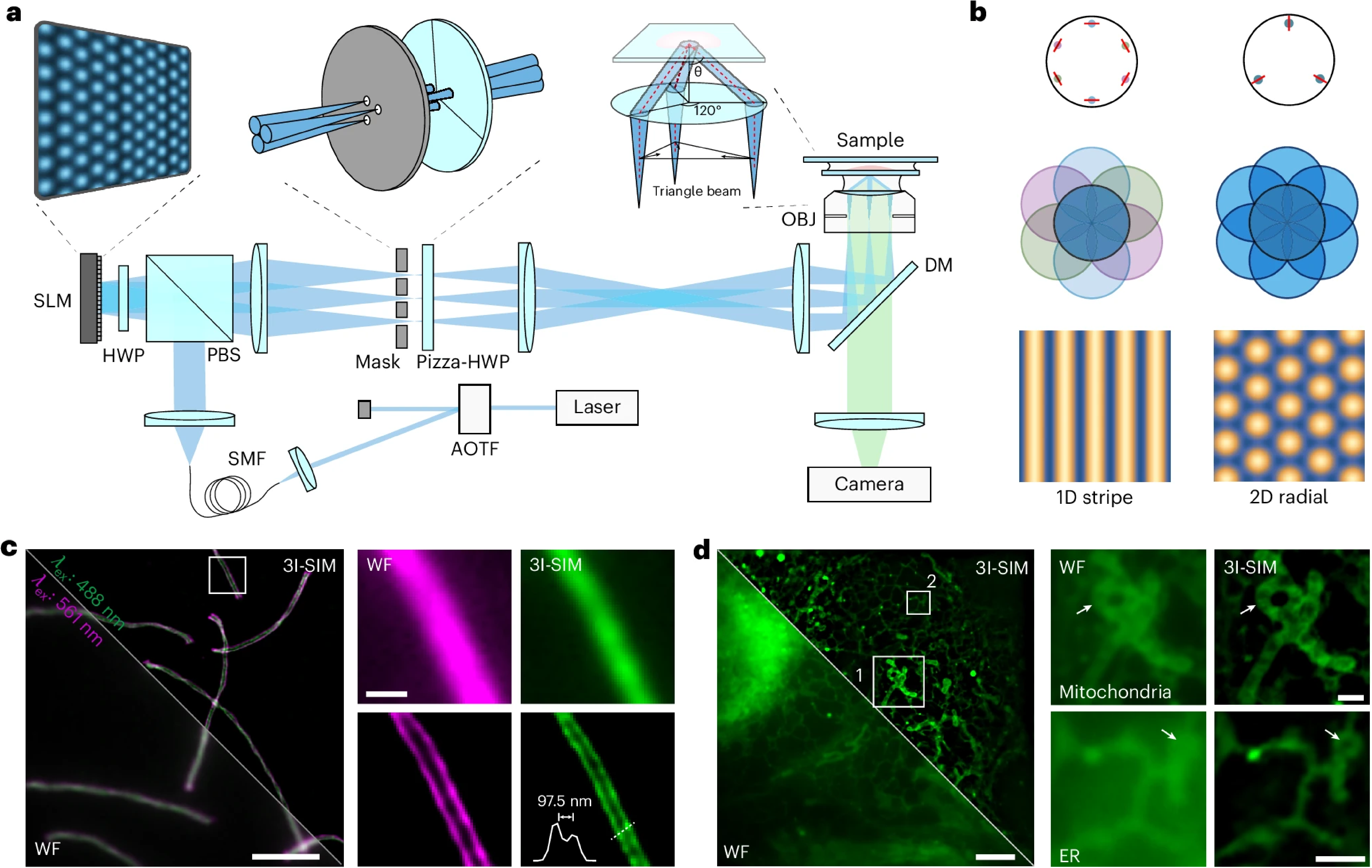
Figure 1. Principle and performance characterization of 3I-SIM. a. Schematic of the 3I-SIM imaging system. b. Comparison of conventional two-beam interference modulation and 3I-SIM triangular-beam interference modulation patterns. c. Dual-color imaging of synaptonemal complexes, comparing widefield and super-resolution results. d. Imaging of Nile Red–labeled COS-7 cells, comparing widefield and super-resolution results.
<関連情報>
- https://newsen.pku.edu.cn/news_events/news/research/15060.html
- https://www.nature.com/articles/s41566-025-01730-0
三角ビーム干渉構造照明顕微鏡法 Triangle-beam interference structured illumination microscopy
Yunzhe Fu,Yiwei Hou,Qianxi Liang,Wenyi Wang,Xin Chen,Boya Jin,Jing Ling,Qiuchen Gu,Donghyun Kim,Pengli Zheng,Meiqi Li & Peng Xi
Nature Photonics Published:15 August 2025
DOI:https://doi.org/10.1038/s41566-025-01730-0
Abstract
Structured illumination microscopy (SIM) is a powerful tool for live-cell super-resolution imaging. Conventional two-dimensional (2D)-SIM uses one-dimensional stripe patterns and rotates them at three angles to achieve uniform resolution. Here, to alleviate photobleaching and improve the temporal resolution of 2D-SIM, we develop triangle-beam interference SIM (3I-SIM), which generates a 2D lattice pattern based on radially polarized beam interference. The radial polarization enhances the signal-to-noise ratio of the high-frequency components. Compared with conventional 2D-SIM, 3I-SIM reduces photobleaching and improves the temporal resolution to 242 Hz. Benefiting from unidirectional phase shift, 3I-SIM provides threefold higher rolling frame rate than conventional 2D-SIM to visualize fast biological dynamics. We further developed 3I-Net, a deep neural network with a co-supervised training scheme, to enhance the performance of 3I-SIM under an extremely low signal intensity. Its higher sensitivity enables the consecutive acquisition of over 100,000 time points at a spatial resolution of 100 nm. We continuously monitor the fine morphological changes in neuronal growth cones for up to 13 h, as well as the transient signals from actin filaments regulating endoplasmic reticulum dynamics. We believe 3I-SIM will offer a suitable platform to study complex and rapid biological processes with high data throughput.