2025-05-20 ロックフェラー大学
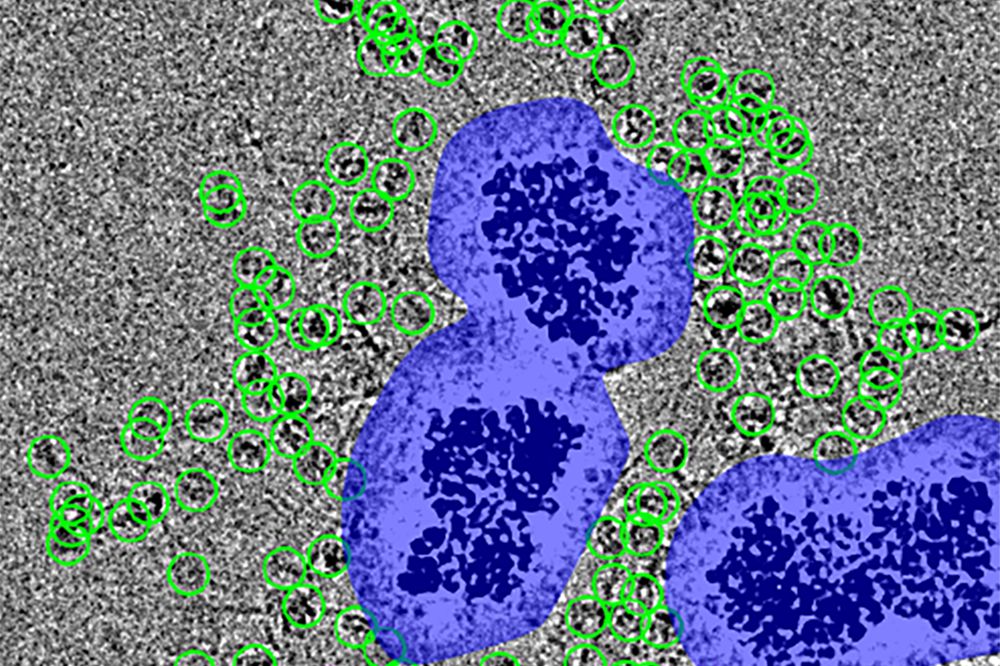 Particles (encircled in green) are spaced at a distance from the noise surrounding the nanobeads (purple), which would otherwise obscure them. (Courtesy of Funabiki Lab)
Particles (encircled in green) are spaced at a distance from the noise surrounding the nanobeads (purple), which would otherwise obscure them. (Courtesy of Funabiki Lab)
<関連情報>
- https://www.rockefeller.edu/news/37486-new-method-dramatically-improves-cryo-ems-imaging-capabilities/
- https://elifesciences.org/articles/103486
MagIC-Cryo-EM、異種試料中の希少高分子の磁気ビーズ上での構造決定 MagIC-Cryo-EM, structural determination on magnetic beads for scarce macromolecules in heterogeneous samples
Yasuhiro Arimura ,Hide A Konishi,Hironori Funabiki
eLife Published:May 20, 2025
DOI:https://doi.org/10.7554/eLife.103486.3
Abstract
Cryo-EM single-particle analyses typically require target macromolecule concentration at 0.05~5.0 mg/ml, which is often difficult to achieve. Here, we devise Magnetic Isolation and Concentration (MagIC)-cryo-EM, a technique enabling direct structural analysis of targets captured on magnetic beads, thereby reducing the targets’ concentration requirement to <0.0005 mg/mL. Adapting MagIC-cryo-EM to a Chromatin Immunoprecipitation protocol, we characterized structural variations of the linker histone H1.8-associated nucleosomes that were isolated from interphase and metaphase chromosomes in Xenopus egg extract. Combining Duplicated Selection To Exclude Rubbish particles (DuSTER), a particle curation method that excludes low signal-to-noise ratio particles, we also resolved the 3D cryo-EM structures of nucleoplasmin NPM2 co-isolated with the linker histone H1.8 and revealed distinct open and closed structural variants. Our study demonstrates the utility of MagIC-cryo-EM for structural analysis of scarce macromolecules in heterogeneous samples and provides structural insights into the cell cycle-regulation of H1.8 association to nucleosomes.