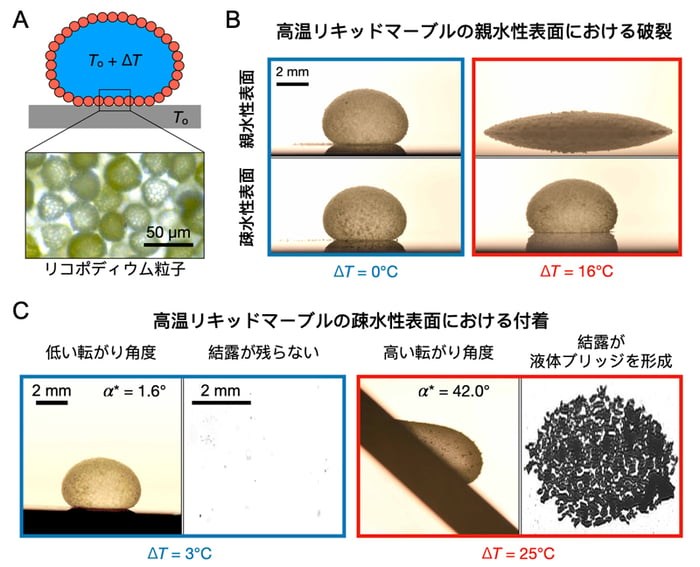
2025-05-12 中国科学院(CAS)
<関連情報>
- https://english.cas.cn/newsroom/research_news/life/202505/t20250513_1043482.shtml
- https://www.sciencedirect.com/science/article/pii/S1055790325000788
アジア産シソ科植物の系統と多様化に関する系統ゲノミクスの新たな知見 Phylogenomics provides new insight into the phylogeny and diversification of Asian Lappula (Boraginaceae)
Dan-Hui Liu, Quan-Ru Liu, Komiljon Sh. Tojibaev, Alexander P. Sukhorukov, Hafiz Muhammad Wariss, Yue Zhao, Lei Yang, Wen-Jun Li
Molecular Phylogenetics and Evolution Available online: 24 April 2025
DOI:https://doi.org/10.1016/j.ympev.2025.108361
Graphical abstract

Highlights
- Integration of 475 nuclear loci and complete plastome sequences for phylogenetic inference within Lappula.
- Polyphyly of Lappula was confirmed.
- Significant gene tree discordance was detected, primarily resulting from incomplete lineage sorting (ILS) and hybridization.
- Hybridization appears to play a crucial role in the evolution and diversification of Lappula.
Abstract
The application of omics data serves as a powerful tool for investigating the roles of incomplete lineage sorting (ILS) and hybridization in shaping genomic diversity, offering deeper insights into complex evolutionary processes. In this study, we utilized deep genome sequencing data from 76 individuals of Lappula and its closely allied genera, collected from China and Central Asia. By employing the HybPiper and Easy353 pipelines, we recovered 262–279 single-copy nuclear genes (SCNs) and 352–353 Angiosperms353 genes, respectively. We analyzed multiple datasets, including complete chloroplast genomes and a filtered set of 475 SCNs, to conduct phylogenetic analyses using both concatenated and coalescent-based methods. Furthermore, we employed Quartet Sampling (QS), coalescent simulations, MSCquartets, HyDe, and reticulate network analyses to investigate the sources of phylogenetic discordance. Our results confirm that Lappula is polyphyletic, with L. mogoltavica clustering with Pseudolappula sinaica and forming a sister relationship with other taxa included in this study. Additionally, three Lepechiniella taxa nested within distinct clades of Lappula. Significant gene tree discordance was observed at several nodes within Lappula. Coalescent simulations and hybrid detection analyses suggest that both ILS and hybridization contribute to these discrepancies. Flow cytometry (FCM) analyses confirmed the presence of both diploid and tetraploid taxa within Lappula. Phylogenetic network analyses further revealed that Clades IV and VII likely originated through hybridization, with the tetraploids in Clade IV arising from two independent hybridization events. Additionally, the “ghost lineage” identified as sister to Lappula redowskii serves as one of the donors in allopolyploidization. In conclusion, our study provides new insights into the deep phylogenetic relationships of Asian Lappula and its closely allied genera, contributing to a more comprehensive understanding of the evolution and diversification of Lappula.