2025-03-05 イリノイ大学アーバナ・シャンペーン校
<関連情報>
- https://aces.illinois.edu/news/studies-reveal-key-genes-corn-architecture-identifying-future-breeding-targets
- https://www.nature.com/articles/s41467-025-56884-w
- https://academic.oup.com/genetics/article-abstract/228/4/iyae162/7832360
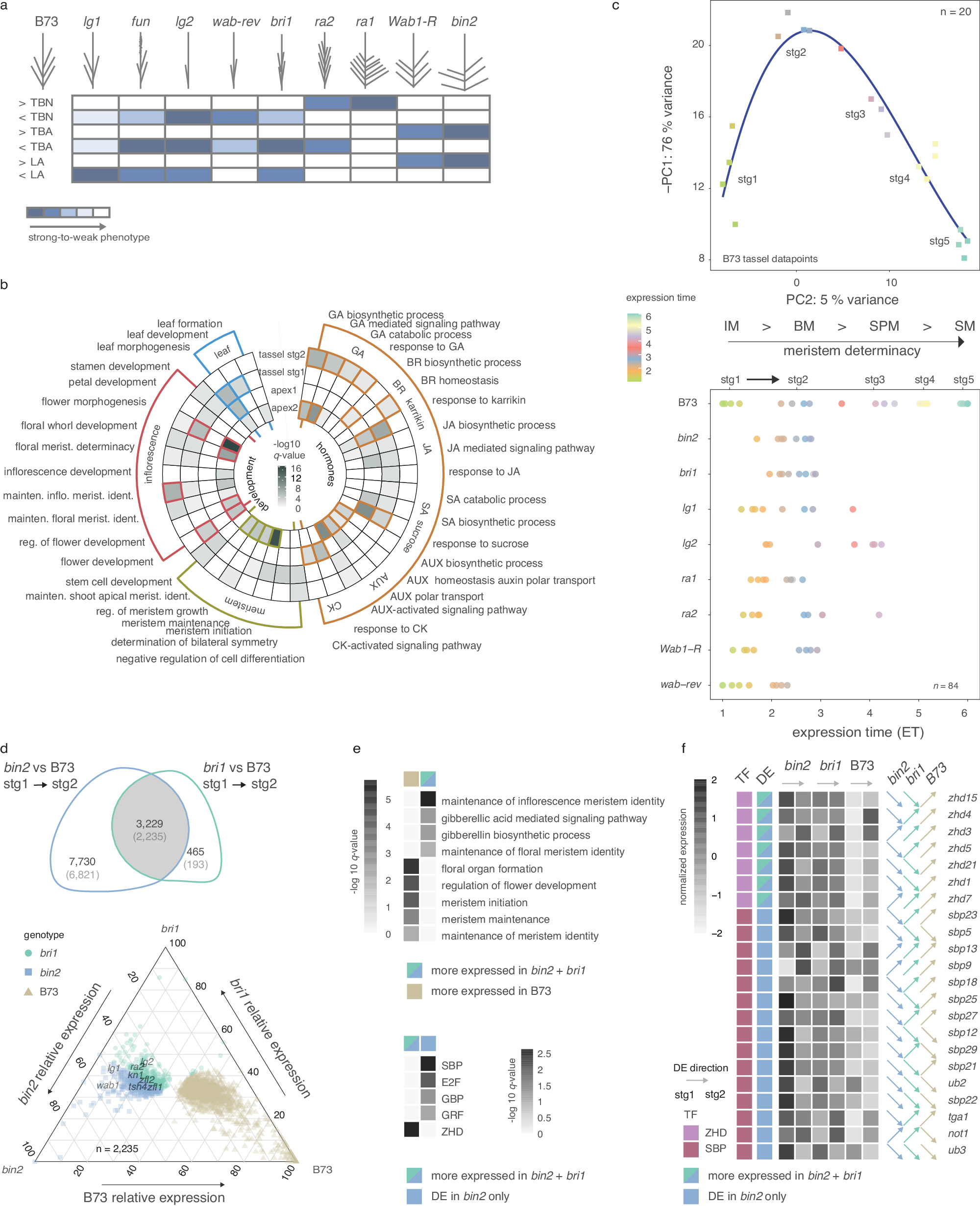
トウモロコシにおける建築的多面性を制御する調節変異 Regulatory variation controlling architectural pleiotropy in maize
Edoardo Bertolini,Brian R. Rice,Max Braud,Jiani Yang,Sarah Hake,Josh Strable,Alexander E. Lipka & Andrea L. Eveland
Nature Communications Published:03 March 2025
DOI:https://doi.org/10.1038/s41467-025-56884-w
Abstract
An early event in plant organogenesis is establishment of a boundary between the stem cell containing meristem and differentiating lateral organ. In maize (Zea mays), evidence suggests a common gene network functions at boundaries of distinct organs and contributes to pleiotropy between leaf angle and tassel branch number, two agronomic traits. To uncover regulatory variation at the nexus of these two traits, we use regulatory network topologies derived from specific developmental contexts to guide multivariate genome-wide association analyses. In addition to defining network plasticity around core pleiotropic loci, we identify new transcription factors that contribute to phenotypic variation in canopy architecture, and structural variation that contributes to cis-regulatory control of pleiotropy between tassel branching and leaf angle across maize diversity. Results demonstrate the power of informing statistical genetics with context-specific developmental networks to pinpoint pleiotropic loci and their cis-regulatory components, which can be used to fine-tune plant architecture for crop improvement.
トウモロコシの遺伝子制御回路に基づくモデルを用いた穀物構造形質のゲノム予測 Genomic prediction of cereal crop architectural traits using models informed by gene regulatory circuitries from maize
Edoardo Bertolini, Mohith Manjunath, Weihao Ge, Matthew D Murphy, Mirai Inaoka, Christina Fliege, Andrea L Eveland, Alexander E Lipka
Genetics Published:23 October 2024
DOI:https://doi.org/10.1093/genetics/iyae162
Abstract
Plant architecture is a major determinant of planting density, which enhances productivity potential for crops per unit area. Genomic prediction is well positioned to expedite genetic gain of plant architectural traits since they are typically highly heritable. Additionally, the adaptation of genomic prediction models to query predictive abilities of markers tagging certain genomic regions could shed light on the genetic architecture of these traits. Here, we leveraged transcriptional networks from a prior study that contextually described developmental progression during tassel and leaf organogenesis in maize (Zea mays) to inform genomic prediction models for architectural traits. Since these developmental processes underlie tassel branching and leaf angle, 2 important agronomic architectural traits, we tested whether genes prioritized from these networks quantitatively contribute to the genetic architecture of these traits. We used genomic prediction models to evaluate the ability of markers in the vicinity of prioritized network genes to predict breeding values of tassel branching and leaf angle traits for 2 diversity panels in maize and diversity panels from sorghum (Sorghum bicolor) and rice (Oryza sativa). Predictive abilities of markers near these prioritized network genes were similar to those using whole-genome marker sets. Notably, markers near highly connected transcription factors from core network motifs in maize yielded predictive abilities that were significantly greater than expected by chance in not only maize but also closely related sorghum. We expect that these highly connected regulators are key drivers of architectural variation that are conserved across closely related cereal crop species.