2022-02-21 オークリッジ国立研究所
Northwestern University and Oak Ridge National Laboratory engineered a microbe, shown in light blue, to convert molecules of industrial waste gases, such as carbon dioxide and carbon monoxide, into acetone. The same microbe can also make isopropanol.
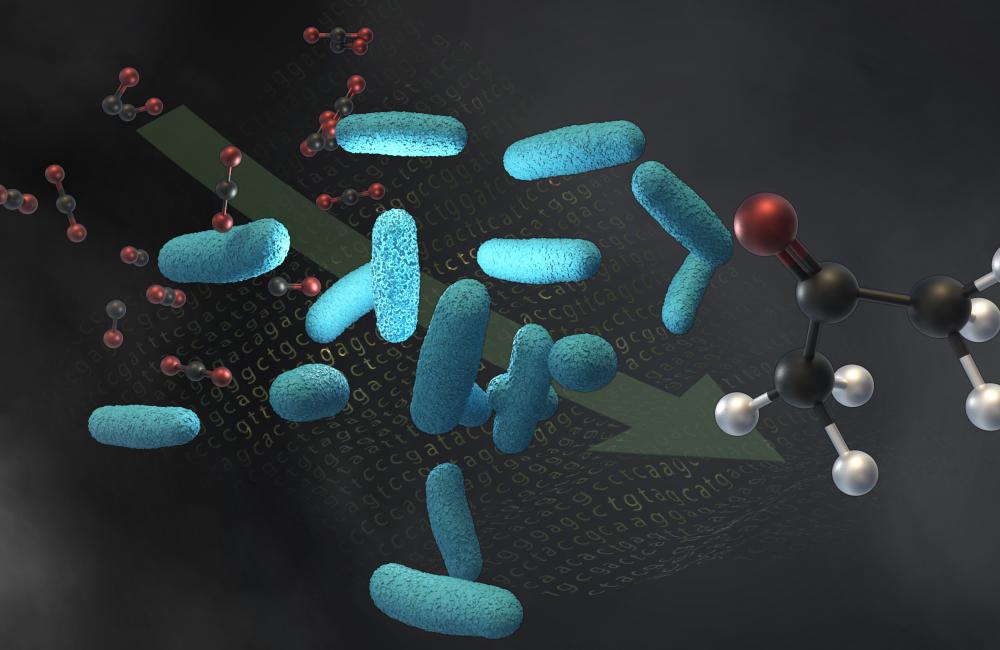
Scientists from LanzaTech, Credit: Andy Sproles/ORNL, U.S. Dept. of Energy
A team of scientists from LanzaTech, Northwestern University and the Department of Energy’s Oak Ridge National Laboratory have developed carbon capture technology that harnesses emissions from industrial processes to produce acetone and isopropanol, known as IPA. These widely used chemicals serve as the basis of thousands of products, from fuels and solvents to acrylic glass and fabrics.
The carbon-negative platform uses microorganisms as tiny but powerful factories that convert carbon from agricultural, industrial and societal waste streams into useful chemicals. The process recycles carbon that would otherwise be released as greenhouse gases that accelerate climate change. In the race to net-zero emissions, this technology provides a step toward a circular carbon economy that can replace products made from fossil resources.
Researchers built on LanzaTech technology to develop an efficient new process that converts waste gases, such as emissions from heavy industry or syngas generated from biomass, into either acetone or IPA, using an engineered bacterium called Clostridium autoethanogenum, or C. auto. Their methods, including a pilot-scale demonstration and life cycle analysis showing the economic viability, are published in the journal Nature Biotechnology.
“This bioprocess provides a sustainable alternative to today’s production routes to these essential chemicals, which currently rely on fresh fossil feedstocks and result in significant toxic waste,” said Jennifer Holmgren, CEO of LanzaTech. “We can reduce greenhouse gases by more than 160%, achieve carbon-negative production and lock up carbon that would have ended up in the atmosphere.”
LanzaTech is currently scaling up this technology, which can be inserted into their existing systems and deployed for use around the world.
“Synthetic biology can be a powerful tool in the quest to advance decarbonization and address climate change,” ORNL Associate Laboratory Director Stan Wullschleger said. “Our scientists leverage world-class capabilities, working closely with industry to harness biological systems to produce valuable fuels and chemicals that support a thriving national bioeconomy.”
The research began at LanzaTech, where scientists previously commercialized a process using C. auto strains that can produce ethanol, a common biofuel, from carbon emissions. Identifying the best enzymes for acetone and IPA production and engineering microbial strains to achieve efficient, high-yielding carbon-to-chemical conversion presented a complex scientific challenge.
The scientists used a three-pronged approach that comprised innovations in pathway screening, strain optimization and process development. As a first step, LanzaTech screened nearly 300 strains for enzymes that could be useful in the acetone and IPA-producing pathways. After identifying useful strains, the scientists built a combinatorial DNA library – the largest ever for this class of microbe – to find enzyme variants that optimized acetone production.
Further optimization relied on cutting-edge synthetic biology tools, including cell-free prototyping by Northwestern University, advanced modeling by LanzaTech and molecular analyses by ORNL.
“Oak Ridge has very unique capabilities in terms of DNA sequencing, systems biology and various metabolomics and proteomics,” Michael Köpke, LanzaTech’s vice president for synthetic biology, said. “Oak Ridge’s expertise helped us troubleshoot the process to find out which steps may be limiting.”
Proteomics, the study of proteins, and metabolomics, the study of small molecules called metabolites, provide a molecular-level view of which specific chemicals are being used and produced by a microbe. Like any organism, when microbes consume or metabolize the substances they need to survive, they produce byproducts. For scientists engineering microbes to produce certain substances, these byproducts represent bottlenecks.
“The protein and metabolite profiles show where a production bottleneck is occurring inside the C. auto cell,” said Tim Tschaplinski, head of ORNL’s Biodesign and Systems Biology Section. “We can see what needs to be modified next in the pathways to flow more of the carbon to the product.”
In this case of C. auto, ORNL scientists determined that the microbe was producing a significant amount of the compound 3-hydroxybutyrate that would require downstream treatment and increase process costs. This compound sits in the middle of a key metabolic pathway where it can move carbon in different directions.
“We look at the enzyme pathways and say, ‘Here’s the block, and there may be a dozen different enzymes within the original collection that may do a better job,’” Tschaplinski said. “Then our partners at Northwestern University express those enzymes in cell-free systems, and we look at what accumulates, which feeds into LanzaTech’s advanced computational models.”
The optimization process was enabled by ORNL’s holistic systems biology approach, which gives scientists a more comprehensive view of what’s happening in the cell and how to improve it, Tschaplinski said. “We use one ‘omic to confirm another,” he said. “By looking at the system as a whole rather than just an individual data stream, we can explore different avenues to enhance the generation of the desired product.”
“We found one of the enzymes, in particular, gave a significant boost once we increased production,” Köpke said. “And we found that through a lot of systems biology and proteomics analyses that were done by Oak Ridge.”
This collaboration is the latest in a longstanding relationship between ORNL and LanzaTech. In 2015, a team of ORNL and LanzaTech scientists sequenced the entire C. auto genome, laying the groundwork for the current research.
The acetone strain and process development, genome-scale modeling, life cycle analysis and initial pilot runs were supported by LanzaTech and the Bioenergy Technologies Office in DOE’s Office of Energy Efficiency and Renewable Energy. The cell-free prototyping and ‘omics analyses were funded by the Biological and Environmental Research program in DOE’s Office of Science. DNA sequencing was supported by the Joint Genome Institute, a DOE Office of Science user facility.
UT-Battelle manages ORNL for the Department of Energy’s Office of Science, the single largest supporter of basic research in the physical sciences in the United States. The Office of Science is working to address some of the most pressing challenges of our time. For more information, please visit energy.gov/science. – Abby Bower and Kim Askey