2025-12-16 中国科学院(CAS)
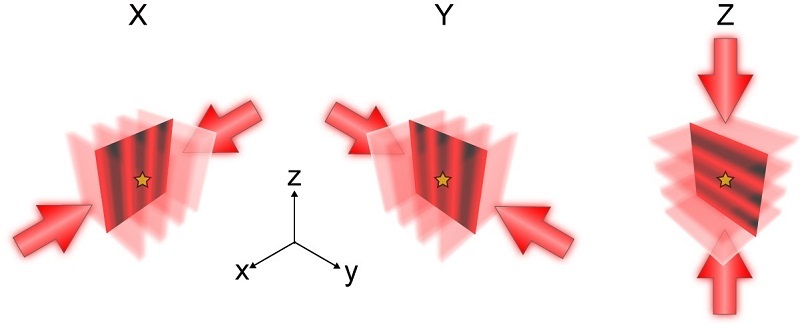
Figure 1. Principle of ROSE-3D (Image by XU Tao’s group and JI Wei’s group)
<関連情報>
- https://english.cas.cn/newsroom/research_news/life/202512/t20251212_1136068.shtml
- https://www.nature.com/articles/s41592-025-02911-z
干渉の局在化による分子スケール等方性3D超解像顕微鏡 Molecular-scale isotropic 3D super-resolution microscopy via interference localization
Shihang Luo (罗世行),Xian’ao Zhao (赵先傲),Yuanyuan Li (李媛媛),Chunyan Fan (樊春燕),Ruina Liu (刘瑞娜),Ran Gong (龚燃),Weixing Li (李尉兴),Nana Ma (马娜娜),Zhenghong Yang (杨郑鸿),Tao Xu (徐涛),Wei Ji (纪伟) & Lusheng Gu (谷陆生)
Nature Methods Published:02 December 2025
DOI:https://doi.org/10.1038/s41592-025-02911-z
Abstract
Three-dimensional (3D) nanoscale imaging reveals the detailed morphology of subcellular structures; however, conventional single-molecule localization microscopy is constrained by limited axial resolution. Here we introduce ROSE-3D, an interferometric localization approach that enables isotropic 3D super-resolution imaging with uniform performance across the entire depth of field. Compared with conventional astigmatism-based methods, ROSE-3D improves lateral localization precision by 2–6 times and axial precision by 3.5–8 times over a depth of field of approximately 1.2 μm. Leveraging its multicolor and whole-cell imaging capabilities, ROSE-3D resolves, in situ, the nanoscale organization of nuclear lamins and the assemblies of mitochondrial fission-related protein DRP1. These results establish ROSE-3D as a powerful tool for interrogating nanoscale cellular architecture.