2025-08-11 マックス・プランク研究所
<関連情報>
- https://www.mpg.de/25177792/genetic-switches-for-adaptability
- https://www.nature.com/articles/s41588-025-02246-7
転写因子結合部位における遺伝的変異は、トウモロコシの表現型遺伝率を主に説明する Genetic variation at transcription factor binding sites largely explains phenotypic heritability in maize
Julia Engelhorn,Samantha J. Snodgrass,Amelie Kok,Arun S. Seetharam,Michael Schneider,Tatjana Kiwit,Ayush Singh,Michael Banf,Duong Thi Hai Doan,Merritt Khaipho-Burch,Daniel E. Runcie,Victor A. Sánchez-Camargo,Rechien Bader,J. Vladimir Torres-Rodriguez,Guangchao Sun,Maike Stam,Fabio Fiorani,Sebastian Beier,James C. Schnable,Hank W. Bass,Matthew B. Hufford,Benjamin Stich,Wolf B. Frommer,Jeffrey Ross-Ibarra & Thomas Hartwig
Nature Genetics Published:11 August 2025
DOI:https://doi.org/10.1038/s41588-025-02246-7
Abstract
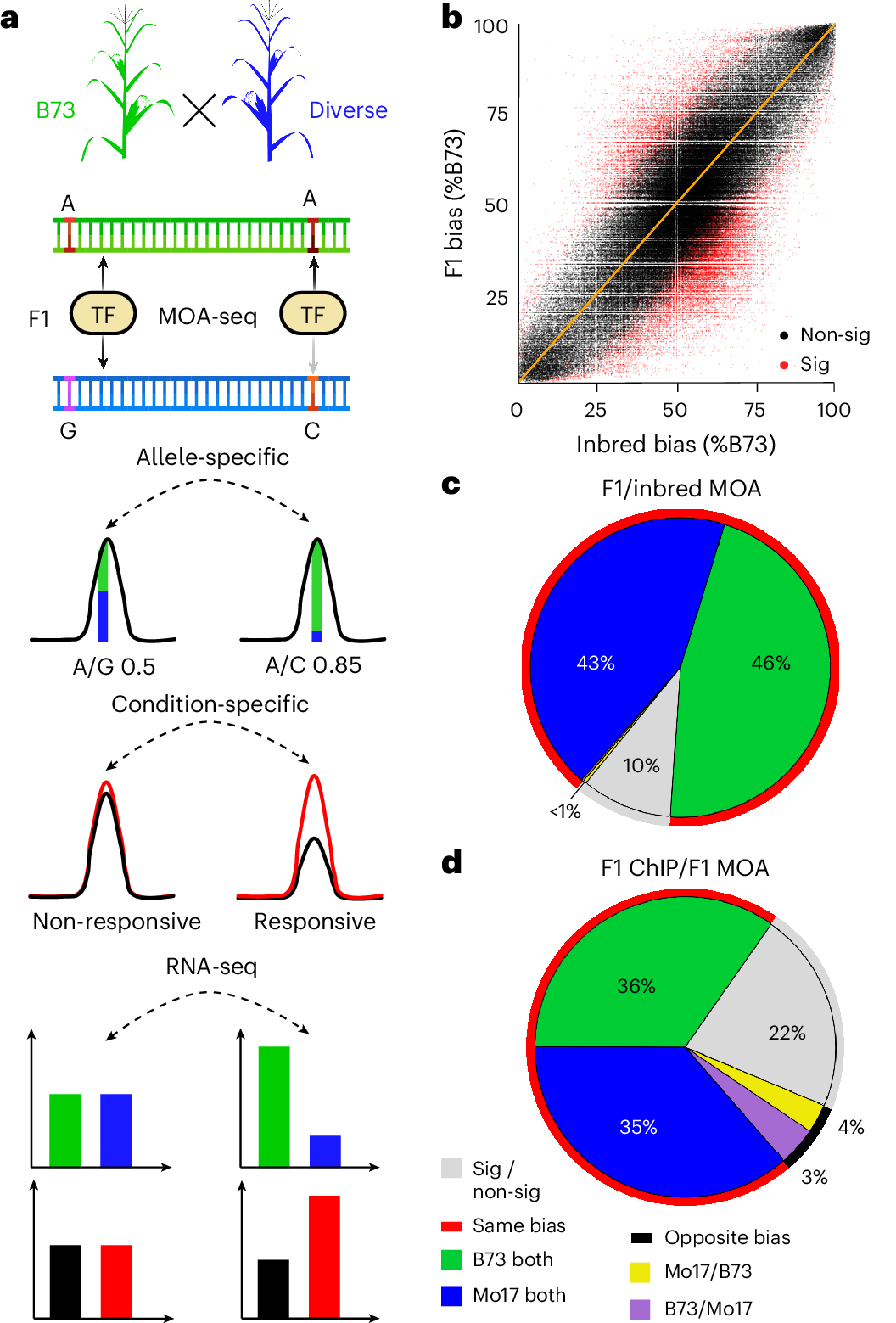
Comprehensive maps of functional variation at transcription factor (TF) binding sites (cis-elements) are crucial for elucidating how genotype shapes phenotype. Here, we report the construction of a pan-cistrome of the maize leaf under well-watered and drought conditions. We quantified haplotype-specific TF footprints across a pan-genome of 25 maize hybrids and mapped over 200,000 variants, genetic, epigenetic, or both (termed binding quantitative trait loci (bQTL)), linked to cis-element occupancy. Three lines of evidence support the functional significance of bQTL: (1) coincidence with causative loci that regulate traits, including vgt1, ZmTRE1 and the MITE transposon near ZmNAC111 under drought; (2) bQTL allelic bias is shared between inbred parents and matches chromatin immunoprecipitation sequencing results; and (3) partitioning genetic variation across genomic regions demonstrates that bQTL capture the majority of heritable trait variation across ~72% of 143 phenotypes. Our study provides an auspicious approach to make functional cis-variation accessible at scale for genetic studies and targeted engineering of complex traits.