2025-07-09 コロンビア大学
<関連情報>
- https://www.engineering.columbia.edu/about/news/need-new-3d-material-build-it-dna
- https://www.nature.com/articles/s41563-025-02263-1
- https://pubs.acs.org/doi/10.1021/acsnano.4c17408
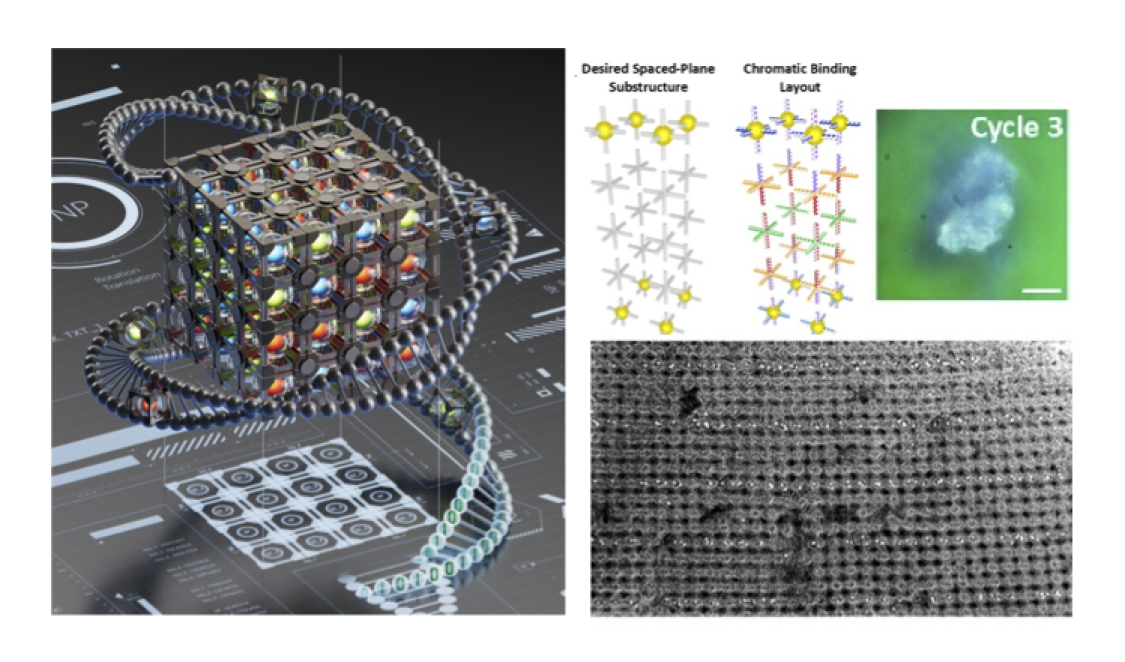
プログラム可能な結合の逆設計による階層的3Dアーキテクチャの符号化 Encoding hierarchical 3D architecture through inverse design of programmable bonds
Jason S. Kahn,Brian Minevich,Aaron Michelson,Hamed Emamy,Jiahao Wu,Huajian Ji,Alexia Yun,Kim Kisslinger,Shuting Xiang,Nanfang Yu,Sanat K. Kumar & Oleg Gang
Nature Materials Published:09 July 2025
DOI:https://doi.org/10.1038/s41563-025-02263-1
Abstract
The ability to fabricate materials and devices at small scales by design has resulted in tremendous technological progress. However, the need for engineered three-dimensional (3D) nanoscale materials requires new strategies for organizing nanocomponents. Here we demonstrate an inverse design approach for the assembly of nanoparticles into hierarchically ordered 3D organizations using DNA voxels with directional, addressable bonds. By identifying intrinsic symmetries in repeating mesoscale structural motifs, we prescribe a set of voxels, termed a mesovoxel, that are assembled into target 3D crystals. The relationship between different degrees of encoded information used for voxel bonds and the fidelity of assembly is investigated using experimental and computational methods. We apply this assembly strategy to create periodic 3D nanoparticle ordered organizations, including structures with low-dimensional elements, helical motifs, a nanoscale analogue of a face-centred perovskite crystal and a distributed Bragg reflector based on a crystal with plasmonic and photonic length-scale regimes.
対称性マッピングによるDNAプログラム可能な3次元結晶の任意設計 Arbitrary Design of DNA-Programmable 3D Crystals through Symmetry Mapping
Jason S. Kahn,Daniel C. Redeker,Aaron Michelson,Alexei Tkachenko,Sarah Hong,Brian Minevich,and Oleg Gang
ACS Nano Published: April 11, 2025
DOI:https://doi.org/10.1021/acsnano.4c17408
Abstract
Nanoscale self-assembly offers exciting potential for creating intricate structures beyond the limits of traditional top-down nanofabrication. Despite advancements in molecularly programmable assembly, particularly utilizing DNA nanotechnology, challenges remain in defining precise assembly instructions for the formation of complex three-dimensional (3D) superlattice architectures. DNA-based self-assembly methods offer programmability through sequence-encoded addressable bonds, but the difficulty lies in reducing the complexity and number of these interactions to establish a modular, structural design strategy and streamline the assembly and component fabrication process. This work proposes a symmetry-mapping bond assignment algorithm to guide the design of arbitrarily prescribed 3D lattices self-assembled from voxels with directional, addressable bonds and capable of carrying nanocargo. The algorithm enables the minimization of the number of DNA-based voxels, thus reducing the amount of information required to encode assembly. The developed approach leverages the symmetries of the target lattices, assembled from voxels, but significantly incorporates experimentally relevant binding rules and restrictions specific to DNA-based systems. We discuss the developed algorithm and demonstrate its capability in selected examples of nanoscale analogs of zinc blende (ZnS) and cubic Laves phase (MgCu2), as well as a lattice based on an arbitrarily designed motif (letter H). Through the established algorithm and associated software for Mapping Of Structurally Encoded aSsembly (MOSES), this inverse design approach provides a scalable solution for designing complexly organized 3D nanostructures, providing a means for programming bottom-up nanomaterial fabrication.