2025-06-27 神戸大学
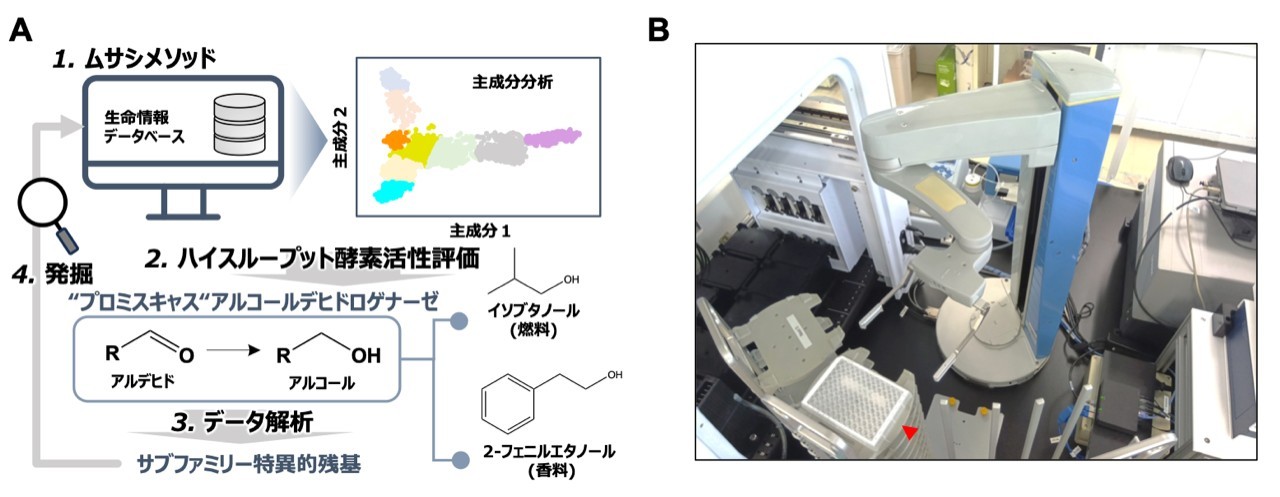
図1(A)有用酵素を発掘するため新規技術の統合型ワークフロー。ムサシメソッドにより、本来の目的とは異なる物質にも働くプロミスキャス酵素の候補を選定し、ハイスループット酵素活性評価システムで酵素の反応速度のデータを収集する。その後、計算科学的手法により、一定の酵素群に共通する配列的特徴(サブファミリー特異的残基)を同定する。これを指標として、有用な酵素を発掘することができる。(B)ハイスループット酵素活性評価のために設計したラボラトリーオートメーション。あらかじめ設置した小型実験プレート(赤やじり)をロボットアームが掴むことから自動実験が開始する。
<関連情報>
- https://www.kobe-u.ac.jp/ja/news/article/20250627-66767/
- https://pubs.acs.org/doi/10.1021/acscatal.5c02764
高活性かつプロミスキャスなアルコール脱水素酵素におけるサブファミリー特異的残基の同定 Identification of Subfamily Specific Residues within Highly Active and Promiscuous Alcohol Dehydrogenases
Ryota Hidese,Kanae Sakai,Musashi Takenaka,Keiji Fushimi,Hisashi Kudo,Kenya Tanaka,Ryo Nasuno,Christopher J. Vavricka,Akihiko Kondo,and Tomohisa Hasunuma
ACS Catalysis Published: June 26, 2025
DOI:https://doi.org/10.1021/acscatal.5c02764
Abstract
Enzyme selection is an essential process in the biobased production of chemicals. It is essential to develop a method to extract yet unknown useful enzymes from protein databases. Enzymes that exhibit substrate promiscuity and high activity hold the potential to access unknown reactions and mediate known reactions with a higher performance. Herein, we propose and validate a principal component analysis (PCA)-based classification method, termed MUSASHI (MUltiple-Sequence Alignment-based protein Selection via clustering using HIgh-dimensional analysis), to identify subfamily-specific residues that are highly conserved among promiscuous alcohol dehydrogenase (ADH). Specifically, zinc-dependent ADH homologues retrieved from the protein database were classified into 9 groups, and according to PCA-based clustering, the activities of 18 ADHs, with representative enzymes from each group, were characterized. As a result, we identified two promiscuous ADH groups: Group 1 ADH, efficient with short-chain and aromatic aldehydes, and Group 3 ADH, efficient with aliphatic and aromatic ketones. Sequence feature analysis then revealed subfamily-specific residues, which are highly conserved only in promiscuous ADH Groups 1 and 3, with the potential to biosynthesize a wide spectrum of target compounds. Tatumella ptyseos ADH, identified from Group 1 of this study, showed higher isobutanol and 2-phenylethanol bioconversions than that of a conventional ADH (Ahr). These results indicate that the MUSASHI method for subfamily-specific residue identification can enable optimal enzyme selection from protein databases.



