2025-05-28 清華大学
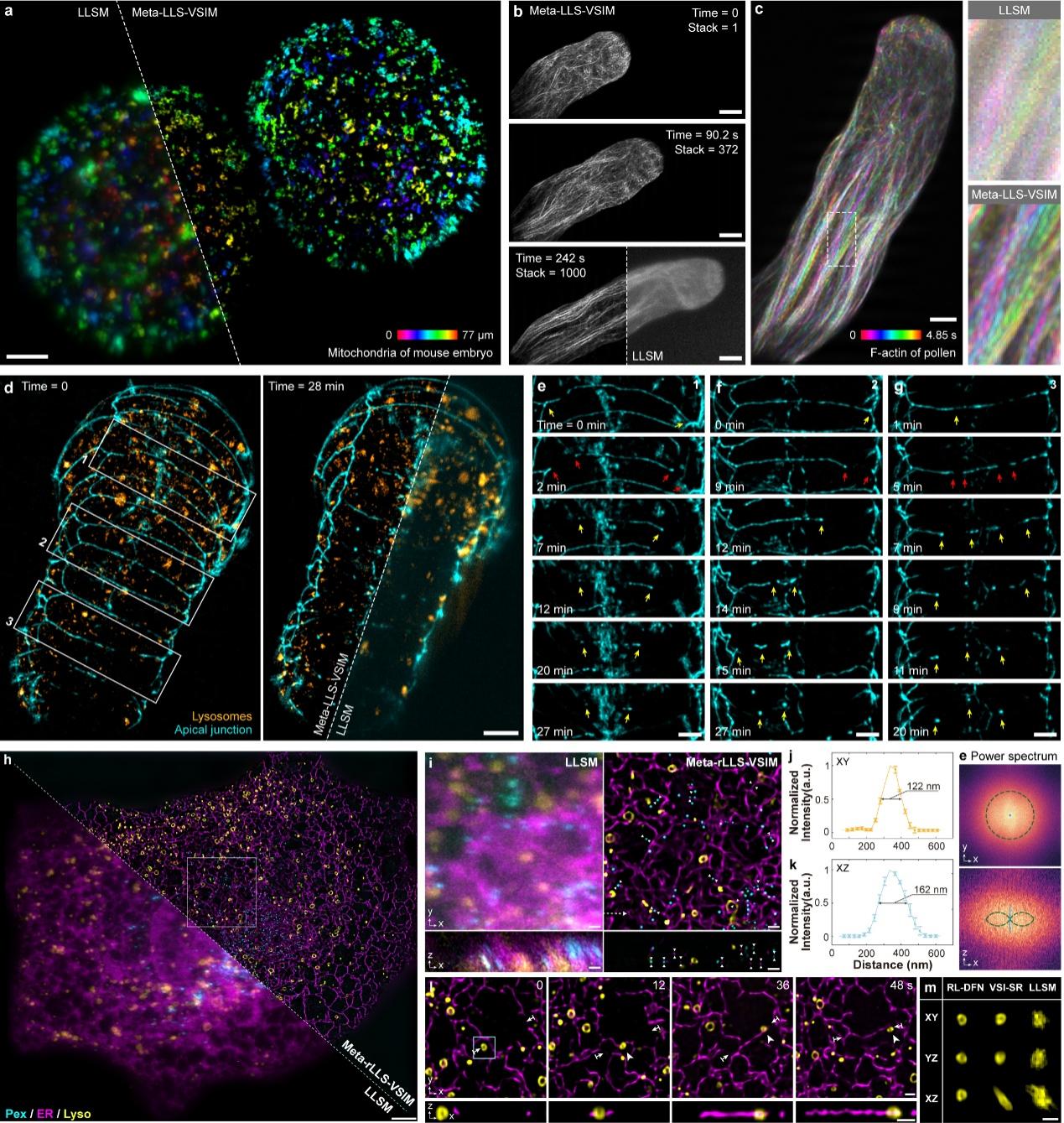 Fig. 1 Demonstration of rapid five-dimensional super-resolution live-cell imaging using Meta-rLLS-VSIM
Fig. 1 Demonstration of rapid five-dimensional super-resolution live-cell imaging using Meta-rLLS-VSIM
<関連情報>
- https://www.tsinghua.edu.cn/en/info/1245/14274.htm
- https://www.nature.com/articles/s41592-025-02678-3
高速・長時間・ほぼ等方的な細胞内イメージングのための高速適応型超解像格子ライトシート顕微鏡法 Fast-adaptive super-resolution lattice light-sheet microscopy for rapid, long-term, near-isotropic subcellular imaging
Chang Qiao,Ziwei Li,Zongfa Wang,Yuhuan Lin,Chong Liu,Siwei Zhang,Yong Liu,Yun Feng,Xiaoyu Yang,Wenfeng Fu,Xue Dong,Jiabao Guo,Wencong Xu,Xinyu Wang,Tao Jiang,Quan Meng,Qinghua Wang,Qionghai Dai & Dong Li
Nature Methods Published:29 April 2025
DOI:https://doi.org/10.1038/s41592-025-02678-3
Abstract
Lattice light-sheet microscopy provides a crucial observation window into intra- and intercellular physiology of living specimens but at the diffraction-limited resolution or anisotropic super-resolution with structured illumination. Here we present meta-learning-empowered reflective lattice light-sheet virtual structured illumination microscopy (Meta-rLLS-VSIM), which upgrades lattice light-sheet microscopy to a near-isotropic super resolution of ~120 nm laterally and ~160 nm axially without modifications of the core optical system or loss of other live-cell imaging metrics. Moreover, we devised an adaptive online training approach by synergizing the front-end imaging system and back-end meta-learning framework, which alleviated the demand for training data by tenfold and reduced the total time for data acquisition and model training down to tens of seconds. We demonstrate the versatile functionalities of Meta-rLLS-VSIM by imaging a variety of bioprocesses with ultrahigh spatiotemporal resolution for hundreds of multicolor volumes, delineating the nanoscale distributions, dynamics and interaction patterns of multiple organelles in embryos and eukaryotic cells.