2025-05-12 カリフォルニア大学リバーサイド校 (UCR)
<関連情報>
- https://news.ucr.edu/articles/2025/05/12/new-computer-language-helps-spot-hidden-pollutants
- https://www.nature.com/articles/s41592-025-02660-z
質量分析データのパターンを見つけるための世界共通言語 A universal language for finding mass spectrometry data patterns
Tito Damiani,Alan K. Jarmusch,Allegra T. Aron,Daniel Petras,Vanessa V. Phelan,Haoqi Nina Zhao,Wout Bittremieux,Deepa D. Acharya,Mohammed M. A. Ahmed,Anelize Bauermeister,Matthew J. Bertin,Paul D. Boudreau,Ricardo M. Borges,Benjamin P. Bowen,Christopher J. Brown,Fernanda O. Chagas,Kenneth D. Clevenger,Mario S. P. Correia,William J. Crandall,Max Crüsemann,Eoin Fahy,Oliver Fiehn,Neha Garg,William H. Gerwick,… Mingxun Wang
Nature Methods Published:12 May 2025
DOI:https://doi.org/10.1038/s41592-025-02660-z
Abstract
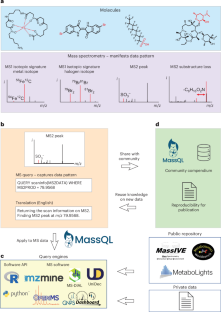
Despite being information rich, the vast majority of untargeted mass spectrometry data are underutilized; most analytes are not used for downstream interpretation or reanalysis after publication. The inability to dive into these rich raw mass spectrometry datasets is due to the limited flexibility and scalability of existing software tools. Here we introduce a new language, the Mass Spectrometry Query Language (MassQL), and an accompanying software ecosystem that addresses these issues by enabling the community to directly query mass spectrometry data with an expressive set of user-defined mass spectrometry patterns. Illustrated by real-world examples, MassQL provides a data-driven definition of chemical diversity by enabling the reanalysis of all public untargeted metabolomics data, empowering scientists across many disciplines to make new discoveries. MassQL has been widely implemented in multiple open-source and commercial mass spectrometry analysis tools, which enhances the ability, interoperability and reproducibility of mining of mass spectrometry data for the research community.