2025-04-16 ミュンヘン大学(LMU)
<関連情報>
- https://www.lmu.de/en/newsroom/news-overview/news/european-potato-genome-small-gene-pool-with-large-differences.html
- https://www.nature.com/articles/s41586-025-08843-0
4倍体ヨーロッパジャガイモの段階的汎ゲノム解析 The phased pan-genome of tetraploid European potato
Hequan Sun,Sergio Tusso,Craig I. Dent,Manish Goel,Raúl Y. Wijfjes,Lisa C. Baus,Xiao Dong,José A. Campoy,Ana Kurdadze,Birgit Walkemeier,Christine Sänger,Bruno Huettel,Ronald C. B. Hutten,Herman J. van Eck,Klaus J. Dehmer & Korbinian Schneeberger
Nature Published:16 April 2025
DOI:https://doi.org/10.1038/s41586-025-08843-0
Abstract
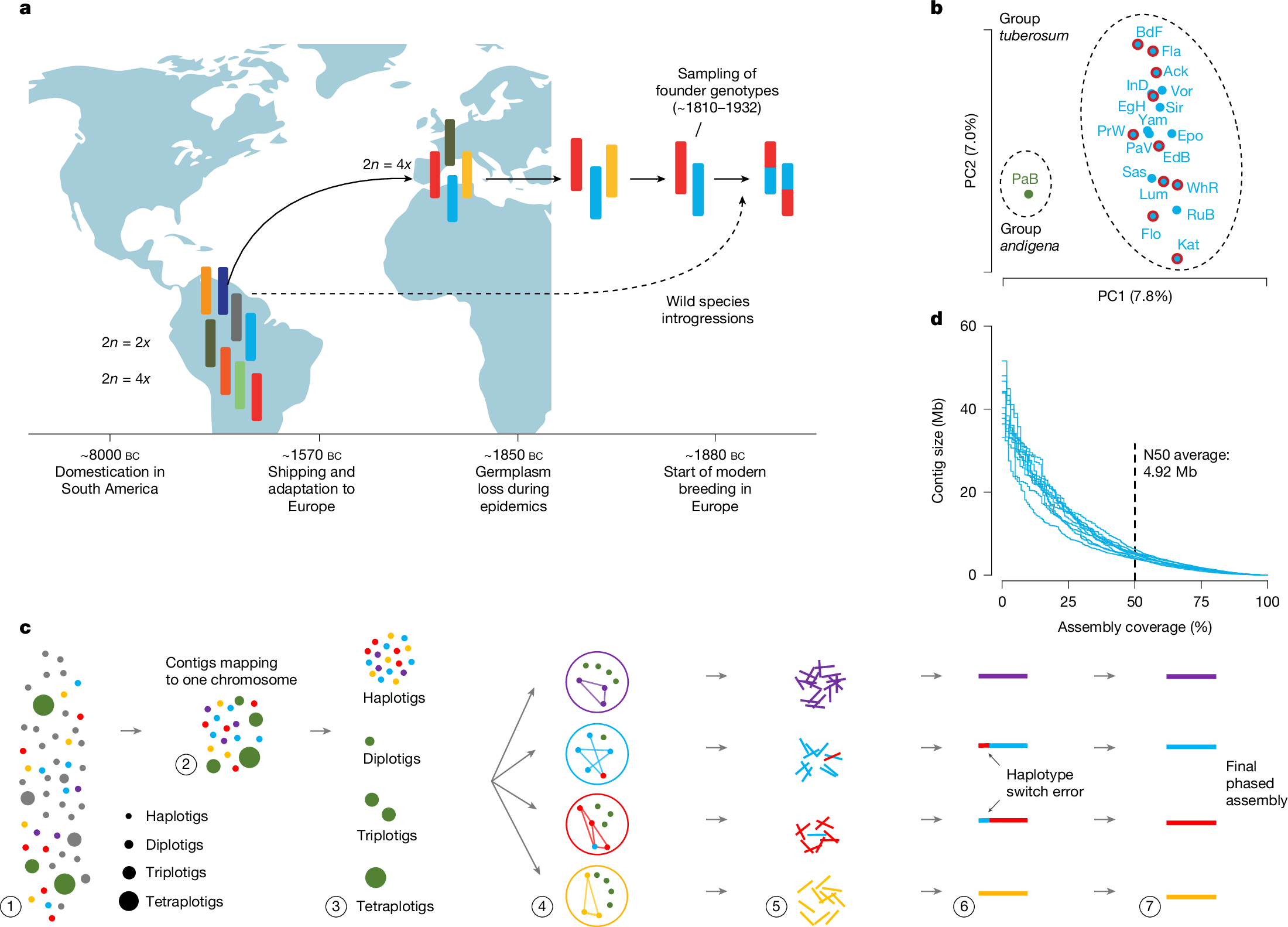
Potatoes were first brought to Europe in the sixteenth century1,2. Two hundred years later, one of the species had become one of the most important food sources across the entire continent and, later, even the entire world3. However, its highly heterozygous, autotetraploid genome has complicated its improvement since then4,5,6,7. Here we present the pan-genome of European potatoes generated from phased genome assemblies of ten historical potato cultivars, which includes approximately 85% of all haplotypes segregating in Europe. Sequence diversity between the haplotypes was extremely high (for example, 20× higher than in humans), owing to numerous introgressions from wild potato species. By contrast, haplotype diversity was very low, in agreement with the population bottlenecks caused by domestication and transition to Europe. To illustrate a practical application of the pan-genome, we converted it into a haplotype graph and used it to generate phased, megabase-scale pseudo-genome assemblies of commercial potatoes (including the famous French fries potato ‘Russet Burbank’) using cost-efficient short reads only. In summary, we present a nearly complete pan-genome of autotetraploid European potato, we describe extraordinarily high sequence diversity in a domesticated crop, and we outline how this resource might be used to accelerate genomics-assisted breeding and research.