2023-06-19 スイス連邦工科大学ローザンヌ校(EPFL)
◆この新しい手法はDNAだけでなく、タンパク質の構成要素であるペプチドなど他の分子にも応用可能であり、医学研究や診断技術の向上に大きな可能性をもたらすと期待されています。
<関連情報>
- https://actu.epfl.ch/news/researchers-control-individual-molecules-for-pre-2/
- https://www.nature.com/articles/s41565-023-01412-4
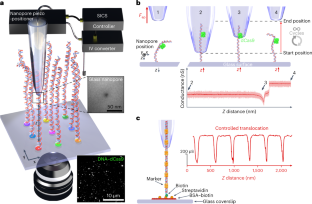
ナノポアを通過する1分子の移動速度を制御して空間的に多重化する Spatially multiplexed single-molecule translocations through a nanopore at controlled speeds
S. M. Leitao,V. Navikas,H. Miljkovic,B. Drake,S. Marion,G. Pistoletti Blanchet,K. Chen,S. F. Mayer,U. F. Keyser,A. Kuhn,G. E. Fantner & A. Radenovic
Nature Nanotechnolog Published:19 June 2023
DOI:https://doi.org/10.1038/s41565-023-01412-4
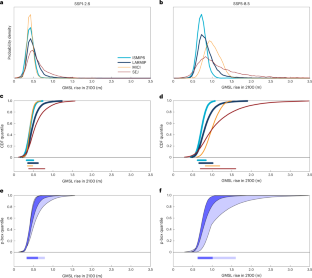
Abstract
In current nanopore-based label-free single-molecule sensing technologies, stochastic processes influence the selection of translocating molecule, translocation rate and translocation velocity. As a result, single-molecule translocations are challenging to control both spatially and temporally. Here we present a method using a glass nanopore mounted on a three-dimensional nanopositioner to spatially select molecules, deterministically tethered on a glass surface, for controlled translocations. By controlling the distance between the nanopore and glass surface, we can actively select the region of interest on the molecule and scan it a controlled number of times and at a controlled velocity. Decreasing the velocity and averaging thousands of consecutive readings of the same molecule increases the signal-to-noise ratio by two orders of magnitude compared with free translocations. We demonstrate the method’s versatility by assessing DNA–protein complexes, DNA rulers and DNA gaps, achieving down to single-nucleotide gap detection.