2025-11-26 ワシントン大学(UW)
<関連情報>
- https://www.washington.edu/news/2025/11/26/counting-salmon-is-a-breeze-with-airborne-edna/
- https://www.nature.com/articles/s41598-025-26293-6
- https://esajournals.onlinelibrary.wiley.com/doi/10.1002/eap.2561
受動的な空気サンプリングにより、水から空気への環境DNAの移動を検出します Passive air sampling detects environmental DNA transfer from water into air
Yin Cheong Aden Ip,Gledis Guri,Elizabeth Andruszkiewicz Allan & Ryan P. Kelly
Scientific Reports Published:26 November 2025
DOI:https://doi.org/10.1038/s41598-025-26293-6
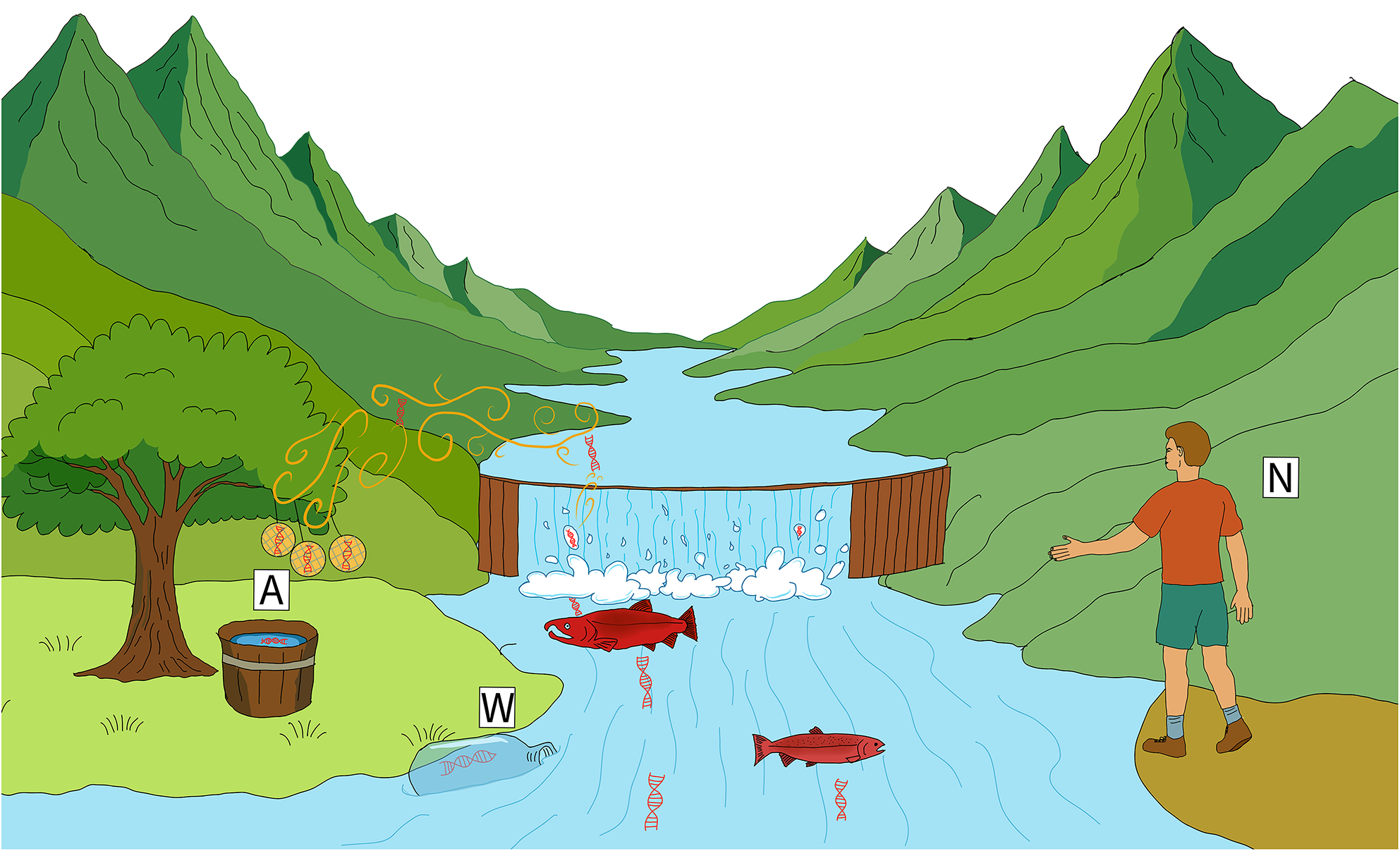
Abstract
Water and air are generally treated as separate reservoirs of environmental DNA (eDNA) derived from the species resident in those respective environmental compartments. However, it is likely that eDNA routinely crosses the air–water boundary in both directions as a result of deposition, evaporation, or other processes. Here, we systematically tested methods of sampling eDNA at the air–water interface, showing for the first time that aquatic life can be consistently detected under standardized field conditions from passive air samples. We deployed four simple air samplers — three different kinds of filters and one open tray of deionized water — alongside paired water samples and visual counts over a six-week peak run of Coho salmon (Oncorhynchus kisutch) at a local spawning stream. We then quantified eDNA concentrations in both air and water (air: copies/cm2/day; water: copies/L) using quantitative PCR, to estimate (1) the concentration of target eDNA in air vs. water, and (2) the capture performance of each filter type. Passive air collectors captured quantitative airborne eDNA signals that covaried with salmon counts, despite air eDNA concentrations being approximately 25,000 times more dilute than water, although eDNA recovery varied with sampler design and orientation. We show the air–water interface can be a quantifiable source of aquatic genetic information in this system using simple, passive samplers that do not require electricity, making them appealing for biomonitoring in remote or resource-limited settings. This work points the way to using airborne eDNA as a promising pathway for biological information critical to conservation, resource management, and public-health protection.
環境DNAで侵略の最前線を追跡 Tracking an invasion front with environmental DNA
Abigail G. Keller, Emily W. Grason, P. Sean McDonald, Ana Ramón-Laca, Ryan P. Kelly
Ecological Applications Published: 06 February 2022
DOI:https://doi.org/10.1002/eap.2561
Abstract
Data from environmental DNA (eDNA) may revolutionize environmental monitoring and management, providing increased detection sensitivity at reduced cost and survey effort. However, eDNA data are rarely used in decision-making contexts, mainly due to uncertainty around (1) data interpretation and (2) whether and how molecular tools dovetail with existing management efforts. We address these challenges by jointly modeling eDNA detection via qPCR and traditional trap data to estimate the density of invasive European green crab (Carcinus maenas), a species for which, historically, baited traps have been used for both detection and control. Our analytical framework simultaneously quantifies uncertainty in both detection methods and provides a robust way of integrating different data streams into management processes. Moreover, the joint model makes clear the marginal information benefit of adding eDNA (or any other) additional data type to an existing monitoring program, offering a path to optimizing sampling efforts for species of management interest. Here, we document green crab eDNA beyond the previously known invasion front and find that the value of eDNA data dramatically increases with low population densities and low traditional sampling effort, as is often the case at leading-edge locations. We also highlight the detection limits of the molecular assay used in this study, as well as scenarios under which eDNA sampling is unlikely to improve existing management efforts.



