2025-04-16 中国科学院(CAS)
<関連情報>
- https://english.cas.cn/newsroom/research_news/life/202504/t20250417_1041391.shtml
- https://www.nature.com/articles/s41586-025-08883-6
野生イネと栽培イネのパンゲノムリファレンス A pangenome reference of wild and cultivated rice
Dongling Guo,Yan Li,Hengyun Lu,Yan Zhao,Nori Kurata,Xinghua Wei,Ahong Wang,Yongchun Wang,Qilin Zhan,Danlin Fan,Congcong Zhou,Yiqi Lu,Qilin Tian,Qijun Weng,Qi Feng,Tao Huang,Lei Zhang,Zhoulin Gu,Changsheng Wang,Ziqun Wang,Zixuan Wang,Xuehui Huang,Qiang Zhao & Bin Han
Nature Published:16 April 2025
DOI:https://doi.org/10.1038/s41586-025-08883-6
Abstract
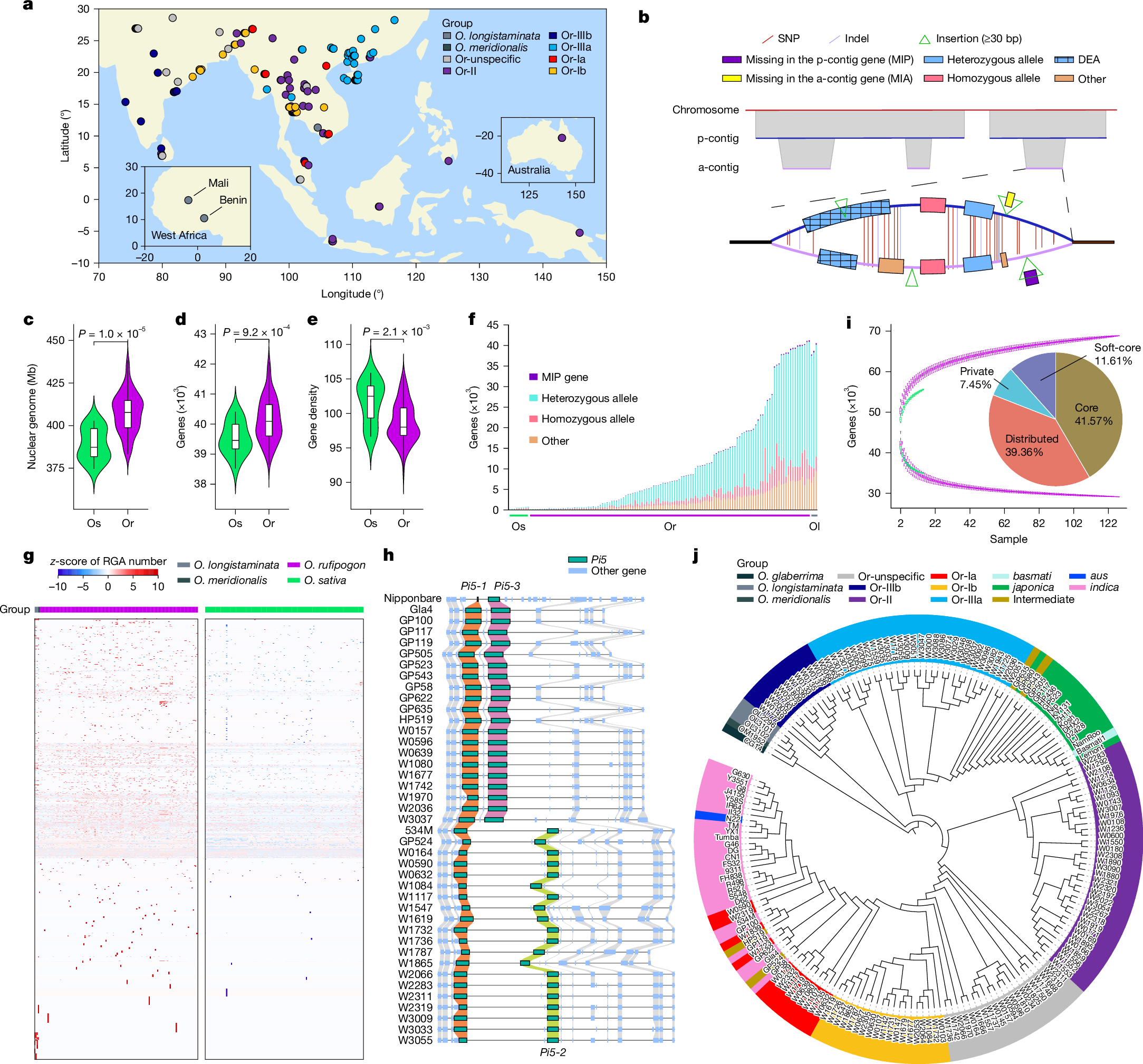
Oryza rufipogon, the wild progenitor of Asian cultivated rice Oryza sativa, is an important resource for rice breeding1. Here we present a wild–cultivated rice pangenome based on 145 chromosome-level assemblies, comprising 129 genetically diverse O. rufipogon accessions and 16 diverse varieties of O. sativa. This pangenome contains 3.87 Gb of sequences that are absent from the O. sativa ssp. japonica cv. Nipponbare reference genome. We captured alternate assemblies that include heterozygous information missing in the primary assemblies, and identified a total of 69,531 pan-genes, with 28,907 core genes and 13,728 wild-rice-specific genes. We observed a higher abundance and a significantly greater diversity of resistance-gene analogues in wild rice than in cultivars. Our analysis indicates that two cultivated subpopulations, intro-indica and basmati, were generated through gene flows among cultivars in South Asia. We also provide strong evidence to support the theory that the initial domestication of all Asian cultivated rice occurred only once. Furthermore, we captured 855,122 differentiated single-nucleotide polymorphisms and 13,853 differentiated presence–absence variations between indica and japonica, which could be traced to the divergence of their respective ancestors and the existence of a larger genetic bottleneck in japonica. This study provides reference resources for enhancing rice breeding, and enriches our understanding of the origins and domestication process of rice.