2024-11-18 オークリッジ国立研究所(ORNL)
<関連情報>
- https://www.ornl.gov/news/targeting-toxic-mercury-prevention
- https://www.pnas.org/doi/10.1073/pnas.2408086121
- https://www.nature.com/articles/s42003-020-1047-5
- https://www.science.org/doi/10.1126/science.1230667
S-アデノシル-L-メチオニンは、膜結合型HgcAB複合体による水銀のメチル化における予想外のメチル供与体である S-adenosyl-L-methionine is the unexpected methyl donor for the methylation of mercury by the membrane-associated HgcAB complex
Kaiyuan Zheng, Katherine W. Rush, Swapneeta S. Date, +5, and Stephen W. Ragsdale
:Proceedings of the National Academy of Sciences Published:November 15, 2024
DOI:https://doi.org/10.1073/pnas.2408086121

Significance
The microbial conversion of inorganic mercury to highly neurotoxic methylmercury is a global environmental concern and a threat to human health. Most mercury methylation research is focused on genomic and microbial aspects, and biochemical studies have been challenging because of the difficulties in expressing and purifying the proteins involved in mercury methylation (HgcAB). The present study establishes the active expression, purification, and characterization of HgcAB as a B12/iron–sulfur enzyme that catalyzes S-adenosyl methionine (SAM)-dependent Hg methylation through a methyl-Co-thiolate intermediate. This work identifies a metabolic role for SAM in heavy metal–associated biological processes.
Abstract
Mercury (Hg) is a heavy metal that exhibits high biological toxicity. Monomethylmercury and dimethylmercury are neurotoxins and a significant environmental concern as they bioaccumulate and biomagnify within the aquatic food web. Microbial Hg methylation involves two proteins, HgcA and HgcB. Here, we show that HgcA and HgcB can be heterologously coexpressed, and the HgcAB complex can be purified. We demonstrated that HgcA is a membrane-associated cobalamin-dependent methyltransferase and HgcB is a ferredoxin-like protein containing two [4Fe-4S] clusters. Further, spectroscopic and kinetic results demonstrate that S-adenosyl-L-methionine (SAM) donates the methyl group to Hg in a two-step reaction involving a methylcob(III)alamin intermediate including Co-thiolate ligation from a conserved Cys residue. Our findings uncover a biological role for SAM in microbial Hg methylation.
メタゲノム配列データを用いたHgcAB複合体の構造決定:微生物の水銀メチル化に関する知見 Structure determination of the HgcAB complex using metagenome sequence data: insights into microbial mercury methylation
Connor J. Cooper,Kaiyuan Zheng,Katherine W. Rush,Alexander Johs,Brian C. Sanders,Georgios A. Pavlopoulos,Nikos C. Kyrpides,Mircea Podar,Sergey Ovchinnikov,Stephen W. Ragsdale & Jerry M. Parks
Communications Biology Published:19 June 2020
DOI:https://doi.org/10.1038/s42003-020-1047-5
Abstract
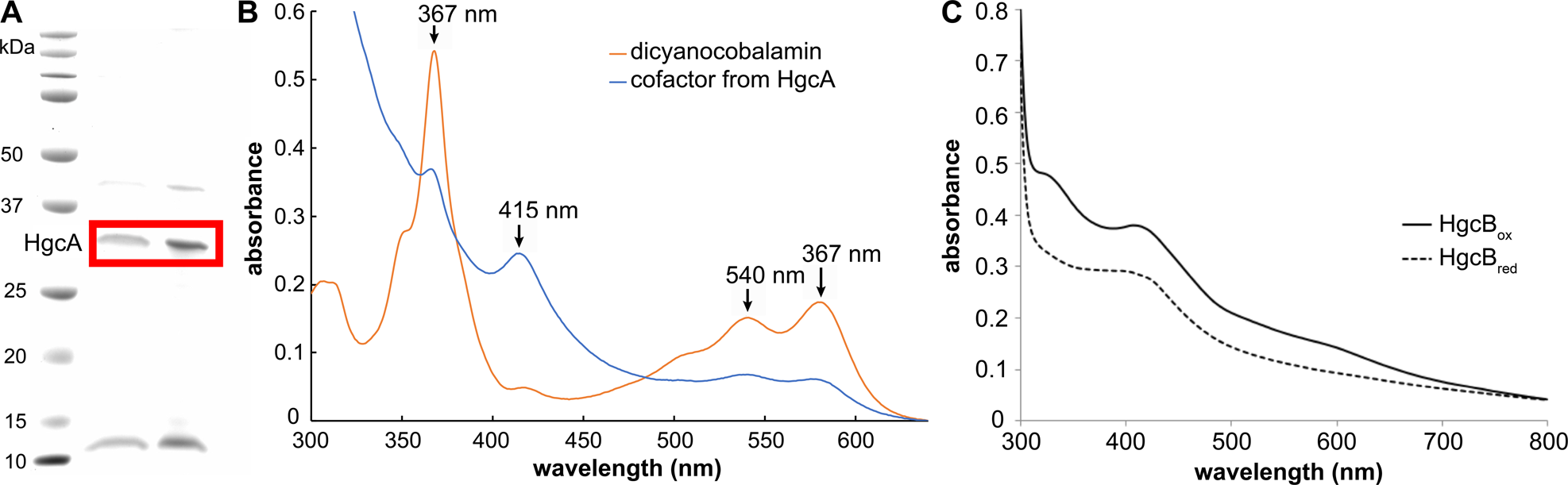
Bacteria and archaea possessing the hgcAB gene pair methylate inorganic mercury (Hg) to form highly toxic methylmercury. HgcA consists of a corrinoid binding domain and a transmembrane domain, and HgcB is a dicluster ferredoxin. However, their detailed structure and function have not been thoroughly characterized. We modeled the HgcAB complex by combining metagenome sequence data mining, coevolution analysis, and Rosetta structure calculations. In addition, we overexpressed HgcA and HgcB in Escherichia coli, confirmed spectroscopically that they bind cobalamin and [4Fe-4S] clusters, respectively, and incorporated these cofactors into the structural model. Surprisingly, the two domains of HgcA do not interact with each other, but HgcB forms extensive contacts with both domains. The model suggests that conserved cysteines in HgcB are involved in shuttling HgII, methylmercury, or both. These findings refine our understanding of the mechanism of Hg methylation and expand the known repertoire of corrinoid methyltransferases in nature.
バクテリアの水銀メチル化の遺伝的基盤 The Genetic Basis for Bacterial Mercury Methylation
Jerry M. Parks, Alexander Johs, Mircea Podar, Romain Bridou, […], and Liyuan Liang
Science Published:7 Feb 2013
DOI:https://doi.org/10.1126/science.1230667
Mercury Methylating Microbes
Mercury (Hg) most commonly becomes bioavailable and enters the food web as the organic form methylmercury, where it induces acute toxicity effects that can be magnified up the food chain. But most natural and anthropogenic Hg exists as inorganic Hg2+ and is only transformed into methylmercury by anaerobic microorganisms—typically sulfur-reducing bacteria. Using comparative genomics, Parks et al. (p. 1332, published online 7 February; see the Perspective by Poulain and Barkay) identified two genes that encode a corrinoid and iron-sulfur proteins in six known Hg-methylating bacteria but were absent in nonmethylating bacteria. In two distantly related model Hg-methylating bacteria, deletion of either gene—or both genes simultaneously—reduced the ability for the bacteria to produce methylmercury but did not impair cellular growth. The presence of this two-gene cluster in several other bacterial and lineages for which genome sequences are available suggests the ability to produce methylmercury may be more broadly distributed in the microbial world than previously recognized.
Abstract
Methylmercury is a potent neurotoxin produced in natural environments from inorganic mercury by anaerobic bacteria. However, until now the genes and proteins involved have remained unidentified. Here, we report a two-gene cluster, hgcA and hgcB, required for mercury methylation by Desulfovibrio desulfuricans ND132 and Geobacter sulfurreducens PCA. In either bacterium, deletion of hgcA, hgcB, or both genes abolishes mercury methylation. The genes encode a putative corrinoid protein, HgcA, and a 2[4Fe-4S] ferredoxin, HgcB, consistent with roles as a methyl carrier and an electron donor required for corrinoid cofactor reduction, respectively. Among bacteria and archaea with sequenced genomes, gene orthologs are present in confirmed methylators but absent in nonmethylators, suggesting a common mercury methylation pathway in all methylating bacteria and archaea sequenced to date.