2024-09-20 カリフォルニア大学リバーサイド校(UCR)
<関連情報>
- https://news.ucr.edu/articles/2024/09/20/new-data-science-tool-greatly-speeds-molecular-analysis-our-environment
- https://www.nature.com/articles/s41596-024-01046-3
非標的メタボロミクスデータから得られた特徴ベース分子ネットワーク結果の統計分析 Statistical analysis of feature-based molecular networking results from non-targeted metabolomics data
Abzer K. Pakkir Shah,Axel Walter,Filip Ottosson,Francesco Russo,Marcelo Navarro-Diaz,Judith Boldt,Jarmo-Charles J. Kalinski,Eftychia Eva Kontou,James Elofson,Alexandros Polyzois,Carolina González-Marín,Shane Farrell,Marie R. Aggerbeck,Thapanee Pruksatrakul,Nathan Chan,Yunshu Wang,Magdalena Pöchhacker,Corinna Brungs,Beatriz Cámara,Andrés Mauricio Caraballo-Rodríguez,Andres Cumsille,Fernanda de Oliveira,Kai Dührkop,Yasin El Abiead,… Daniel Petras
Nature Protocols Published:20 September 2024
DOI:https://doi.org/10.1038/s41596-024-01046-3
Abstract
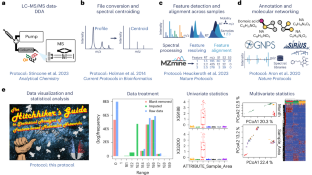
Feature-based molecular networking (FBMN) is a popular analysis approach for liquid chromatography–tandem mass spectrometry-based non-targeted metabolomics data. While processing liquid chromatography–tandem mass spectrometry data through FBMN is fairly streamlined, downstream data handling and statistical interrogation are often a key bottleneck. Especially users new to statistical analysis struggle to effectively handle and analyze complex data matrices. Here we provide a comprehensive guide for the statistical analysis of FBMN results, focusing on the downstream analysis of the FBMN output table. We explain the data structure and principles of data cleanup and normalization, as well as uni- and multivariate statistical analysis of FBMN results. We provide explanations and code in two scripting languages (R and Python) as well as the QIIME2 framework for all protocol steps, from data clean-up to statistical analysis. All code is shared in the form of Jupyter Notebooks (https://github.com/Functional-Metabolomics-Lab/FBMN-STATS). Additionally, the protocol is accompanied by a web application with a graphical user interface (https://fbmn-statsguide.gnps2.org/) to lower the barrier of entry for new users and for educational purposes. Finally, we also show users how to integrate their statistical results into the molecular network using the Cytoscape visualization tool. Throughout the protocol, we use a previously published environmental metabolomics dataset for demonstration purposes. Together, the protocol, code and web application provide a complete guide and toolbox for FBMN data integration, cleanup and advanced statistical analysis, enabling new users to uncover molecular insights from their non-targeted metabolomics data. Our protocol is tailored for the seamless analysis of FBMN results from Global Natural Products Social Molecular Networking and can be easily adapted to other mass spectrometry feature detection, annotation and networking tools.
Key points
- Feature-based molecular networking (FBMN) is a popular workflow for liquid chromatography–tandem mass spectrometry-based non-targeted metabolomics data analysis.
- This protocol provides a detailed guide, code (R, Python and QIIME2) and a web application for FBMN data integration, clean-up and advanced statistical analysis, allowing new and experienced users to uncover molecular insights from their non-targeted metabolomics data.



